
Lignipirellula cremea
Taxonomy: cellular organisms; Bacteria; PVC group; Planctomycetes; Planctomycetia; Pirellulales; Pirellulaceae; Lignipirellula
Average proteome isoelectric point is 6.1
Get precalculated fractions of proteins
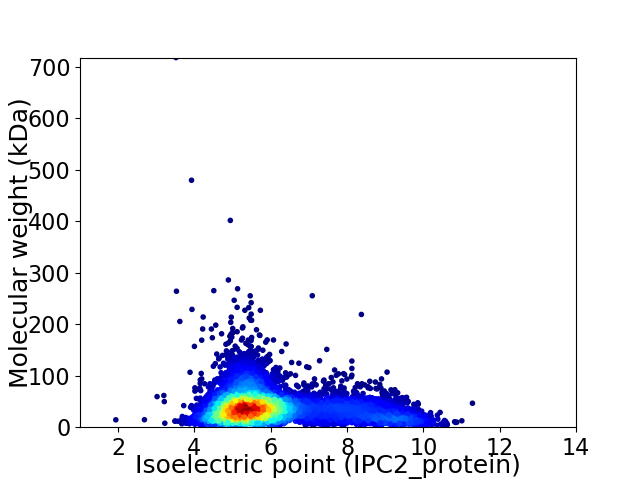
Virtual 2D-PAGE plot for 6917 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A518DQY0|A0A518DQY0_9BACT Adenine phosphoribosyltransferase OS=Lignipirellula cremea OX=2528010 GN=apt PE=3 SV=1
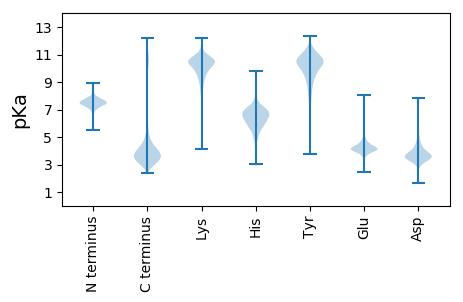
MM1 pKa = 7.59ASSPFDD7 pKa = 3.68NDD9 pKa = 2.64SDD11 pKa = 4.02IPADD15 pKa = 3.56SALDD19 pKa = 3.59MDD21 pKa = 5.31LGGDD25 pKa = 3.67MSLDD29 pKa = 3.42MPVEE33 pKa = 4.11STEE36 pKa = 4.17NAEE39 pKa = 4.07QAPGDD44 pKa = 3.98MFAGVPDD51 pKa = 4.01KK52 pKa = 11.38APAGGPAEE60 pKa = 4.27ADD62 pKa = 3.27APEE65 pKa = 4.32EE66 pKa = 4.34VPVKK70 pKa = 9.76RR71 pKa = 11.84TPSDD75 pKa = 3.63VYY77 pKa = 11.23SWLLAAAFICLCTGVLLLYY96 pKa = 10.12MEE98 pKa = 4.54LRR100 pKa = 11.84DD101 pKa = 3.99YY102 pKa = 11.55GSYY105 pKa = 10.39PYY107 pKa = 10.14WKK109 pKa = 8.56TSGSAAVGASGRR121 pKa = 3.8
MM1 pKa = 7.59ASSPFDD7 pKa = 3.68NDD9 pKa = 2.64SDD11 pKa = 4.02IPADD15 pKa = 3.56SALDD19 pKa = 3.59MDD21 pKa = 5.31LGGDD25 pKa = 3.67MSLDD29 pKa = 3.42MPVEE33 pKa = 4.11STEE36 pKa = 4.17NAEE39 pKa = 4.07QAPGDD44 pKa = 3.98MFAGVPDD51 pKa = 4.01KK52 pKa = 11.38APAGGPAEE60 pKa = 4.27ADD62 pKa = 3.27APEE65 pKa = 4.32EE66 pKa = 4.34VPVKK70 pKa = 9.76RR71 pKa = 11.84TPSDD75 pKa = 3.63VYY77 pKa = 11.23SWLLAAAFICLCTGVLLLYY96 pKa = 10.12MEE98 pKa = 4.54LRR100 pKa = 11.84DD101 pKa = 3.99YY102 pKa = 11.55GSYY105 pKa = 10.39PYY107 pKa = 10.14WKK109 pKa = 8.56TSGSAAVGASGRR121 pKa = 3.8
Molecular weight: 12.66 kDa
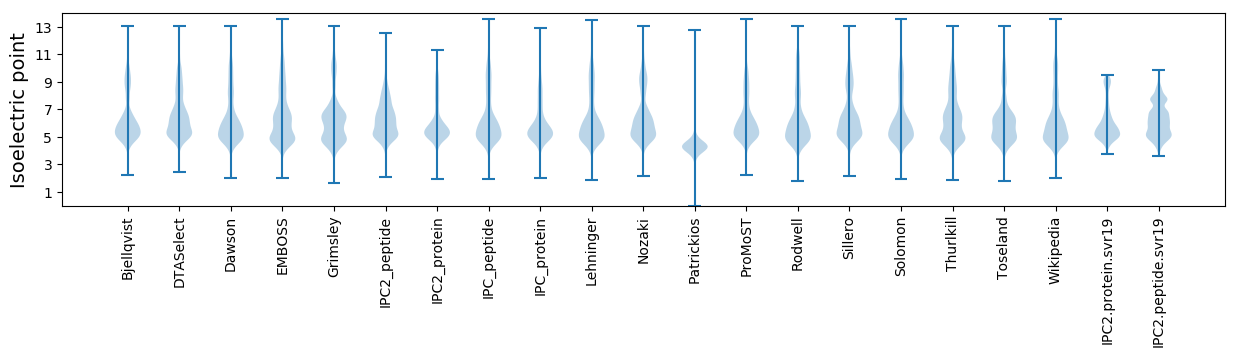
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A518DRY8|A0A518DRY8_9BACT Uncharacterized protein OS=Lignipirellula cremea OX=2528010 GN=Pla8534_23860 PE=4 SV=1
MM1 pKa = 7.18RR2 pKa = 11.84ASWNRR7 pKa = 11.84CVEE10 pKa = 4.28RR11 pKa = 11.84AASPANCRR19 pKa = 11.84PTCVMRR25 pKa = 11.84RR26 pKa = 11.84PIRR29 pKa = 11.84RR30 pKa = 11.84IRR32 pKa = 11.84LVATRR37 pKa = 11.84LPQAPRR43 pKa = 11.84PFRR46 pKa = 11.84ILDD49 pKa = 3.49RR50 pKa = 11.84LAIPLPPPPSRR61 pKa = 11.84LRR63 pKa = 11.84TIRR66 pKa = 11.84SATPLLPLRR75 pKa = 11.84LLLLSLRR82 pKa = 11.84TTIRR86 pKa = 11.84SALLPAILRR95 pKa = 11.84PPRR98 pKa = 11.84LFPRR102 pKa = 11.84TTIRR106 pKa = 11.84SVLLPALPPVTRR118 pKa = 11.84LLPLVMIRR126 pKa = 11.84SAPPPAILRR135 pKa = 11.84PPRR138 pKa = 11.84LAMIRR143 pKa = 11.84SVLLPAILRR152 pKa = 11.84PPRR155 pKa = 11.84LAMIRR160 pKa = 11.84SVHH163 pKa = 6.26PLALPPVTRR172 pKa = 11.84LLPLVMIRR180 pKa = 11.84SVLPPAIPRR189 pKa = 11.84PLLLATIRR197 pKa = 11.84LALLLAIPRR206 pKa = 11.84PPRR209 pKa = 11.84LATIRR214 pKa = 11.84LAHH217 pKa = 7.04LLALLPAPRR226 pKa = 11.84LLRR229 pKa = 11.84LATIRR234 pKa = 11.84SAPPLAILRR243 pKa = 11.84PPRR246 pKa = 11.84LATIRR251 pKa = 11.84LAHH254 pKa = 6.81LPALPPVTRR263 pKa = 11.84LLPLVMIRR271 pKa = 11.84SVLPPAILRR280 pKa = 11.84PLRR283 pKa = 11.84LATIRR288 pKa = 11.84LAHH291 pKa = 7.04LLALLPAPRR300 pKa = 11.84LLRR303 pKa = 11.84LAMIRR308 pKa = 11.84SAPPPAILRR317 pKa = 11.84PLRR320 pKa = 11.84LATIRR325 pKa = 11.84LAHH328 pKa = 7.03LLALLLVTRR337 pKa = 11.84PLRR340 pKa = 11.84LALIRR345 pKa = 11.84SAPLPLLRR353 pKa = 11.84LAPLPLIRR361 pKa = 11.84LAPLPLIRR369 pKa = 11.84SAPLLLLRR377 pKa = 11.84RR378 pKa = 11.84PLPPVGIRR386 pKa = 11.84LAMTRR391 pKa = 11.84LAPIRR396 pKa = 11.84LPRR399 pKa = 11.84LRR401 pKa = 11.84RR402 pKa = 11.84PHH404 pKa = 6.49RR405 pKa = 11.84RR406 pKa = 11.84LPRR409 pKa = 11.84PLLRR413 pKa = 11.84LRR415 pKa = 11.84RR416 pKa = 3.89
MM1 pKa = 7.18RR2 pKa = 11.84ASWNRR7 pKa = 11.84CVEE10 pKa = 4.28RR11 pKa = 11.84AASPANCRR19 pKa = 11.84PTCVMRR25 pKa = 11.84RR26 pKa = 11.84PIRR29 pKa = 11.84RR30 pKa = 11.84IRR32 pKa = 11.84LVATRR37 pKa = 11.84LPQAPRR43 pKa = 11.84PFRR46 pKa = 11.84ILDD49 pKa = 3.49RR50 pKa = 11.84LAIPLPPPPSRR61 pKa = 11.84LRR63 pKa = 11.84TIRR66 pKa = 11.84SATPLLPLRR75 pKa = 11.84LLLLSLRR82 pKa = 11.84TTIRR86 pKa = 11.84SALLPAILRR95 pKa = 11.84PPRR98 pKa = 11.84LFPRR102 pKa = 11.84TTIRR106 pKa = 11.84SVLLPALPPVTRR118 pKa = 11.84LLPLVMIRR126 pKa = 11.84SAPPPAILRR135 pKa = 11.84PPRR138 pKa = 11.84LAMIRR143 pKa = 11.84SVLLPAILRR152 pKa = 11.84PPRR155 pKa = 11.84LAMIRR160 pKa = 11.84SVHH163 pKa = 6.26PLALPPVTRR172 pKa = 11.84LLPLVMIRR180 pKa = 11.84SVLPPAIPRR189 pKa = 11.84PLLLATIRR197 pKa = 11.84LALLLAIPRR206 pKa = 11.84PPRR209 pKa = 11.84LATIRR214 pKa = 11.84LAHH217 pKa = 7.04LLALLPAPRR226 pKa = 11.84LLRR229 pKa = 11.84LATIRR234 pKa = 11.84SAPPLAILRR243 pKa = 11.84PPRR246 pKa = 11.84LATIRR251 pKa = 11.84LAHH254 pKa = 6.81LPALPPVTRR263 pKa = 11.84LLPLVMIRR271 pKa = 11.84SVLPPAILRR280 pKa = 11.84PLRR283 pKa = 11.84LATIRR288 pKa = 11.84LAHH291 pKa = 7.04LLALLPAPRR300 pKa = 11.84LLRR303 pKa = 11.84LAMIRR308 pKa = 11.84SAPPPAILRR317 pKa = 11.84PLRR320 pKa = 11.84LATIRR325 pKa = 11.84LAHH328 pKa = 7.03LLALLLVTRR337 pKa = 11.84PLRR340 pKa = 11.84LALIRR345 pKa = 11.84SAPLPLLRR353 pKa = 11.84LAPLPLIRR361 pKa = 11.84LAPLPLIRR369 pKa = 11.84SAPLLLLRR377 pKa = 11.84RR378 pKa = 11.84PLPPVGIRR386 pKa = 11.84LAMTRR391 pKa = 11.84LAPIRR396 pKa = 11.84LPRR399 pKa = 11.84LRR401 pKa = 11.84RR402 pKa = 11.84PHH404 pKa = 6.49RR405 pKa = 11.84RR406 pKa = 11.84LPRR409 pKa = 11.84PLLRR413 pKa = 11.84LRR415 pKa = 11.84RR416 pKa = 3.89
Molecular weight: 46.58 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2670640 |
29 |
7286 |
386.1 |
42.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
10.893 ± 0.038 | 1.152 ± 0.012 |
5.845 ± 0.023 | 6.291 ± 0.026 |
3.655 ± 0.014 | 7.748 ± 0.03 |
2.208 ± 0.013 | 4.269 ± 0.021 |
3.434 ± 0.023 | 10.472 ± 0.035 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.073 ± 0.013 | 2.845 ± 0.017 |
5.866 ± 0.027 | 4.478 ± 0.025 |
6.9 ± 0.031 | 5.854 ± 0.022 |
5.145 ± 0.023 | 6.814 ± 0.022 |
1.566 ± 0.013 | 2.49 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
