
Murid herpesvirus 4 (MuHV-4) (Murine gammaherpesvirus 68)
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Peploviricota; Herviviricetes; Herpesvirales; Herpesviridae; Gammaherpesvirinae; Rhadinovirus
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
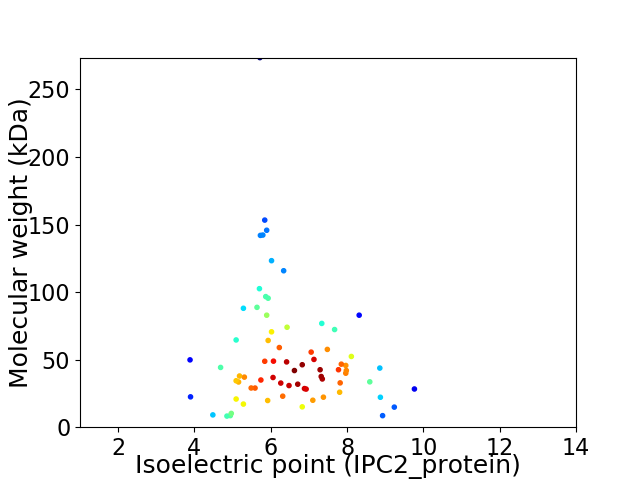
Virtual 2D-PAGE plot for 73 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q98336|Q98336_MHV68 Envelope glycoprotein B OS=Murid herpesvirus 4 OX=33708 GN=8 PE=3 SV=1
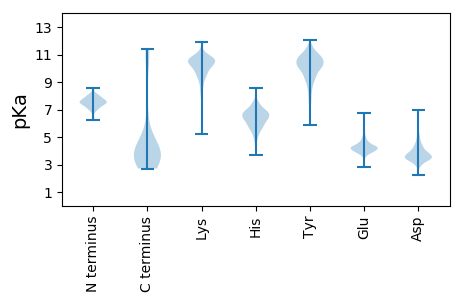
MM1 pKa = 7.58CGVKK5 pKa = 10.29SLAKK9 pKa = 10.34CFLLFQIISFLGNHH23 pKa = 5.98NLVWVPGAALGAAEE37 pKa = 4.32TVEE40 pKa = 5.03GITSRR45 pKa = 11.84EE46 pKa = 3.74MEE48 pKa = 4.23INATKK53 pKa = 10.49APSSGATFSLLVTLSNNNPTTIMRR77 pKa = 11.84PPVAQNGEE85 pKa = 4.54SVHH88 pKa = 6.6KK89 pKa = 10.11DD90 pKa = 2.98ARR92 pKa = 11.84SASASDD98 pKa = 3.6PTTSEE103 pKa = 3.72PTSPGEE109 pKa = 4.34EE110 pKa = 4.11PTEE113 pKa = 4.42ADD115 pKa = 3.46PKK117 pKa = 9.53AAPSAGHH124 pKa = 5.74VGEE127 pKa = 4.99TEE129 pKa = 4.24PEE131 pKa = 4.31SPTPLPATPKK141 pKa = 10.28PSSQEE146 pKa = 4.07DD147 pKa = 4.33NPTMTPPTAEE157 pKa = 4.61PPTSNADD164 pKa = 3.03VSTEE168 pKa = 3.93HH169 pKa = 7.51VDD171 pKa = 3.35EE172 pKa = 5.04TEE174 pKa = 4.32PEE176 pKa = 4.4SPTFLPPTPEE186 pKa = 4.94PDD188 pKa = 3.38TPTTPEE194 pKa = 4.0TTTPSQNQEE203 pKa = 4.18DD204 pKa = 5.16EE205 pKa = 4.49PTLTTSSSDD214 pKa = 3.52APADD218 pKa = 3.79TSDD221 pKa = 3.37TSPPKK226 pKa = 10.48QEE228 pKa = 4.89DD229 pKa = 4.1DD230 pKa = 3.6PVKK233 pKa = 9.38PTEE236 pKa = 4.54SKK238 pKa = 10.39PQAEE242 pKa = 4.78PKK244 pKa = 10.41DD245 pKa = 3.93NSPSDD250 pKa = 3.81VPEE253 pKa = 4.39TADD256 pKa = 4.38SPTDD260 pKa = 3.51PASPTVEE267 pKa = 4.05LTPPTEE273 pKa = 4.31PPTPEE278 pKa = 4.09TVSPADD284 pKa = 3.54SPVPQPTAPAEE295 pKa = 4.03PSKK298 pKa = 10.77PEE300 pKa = 3.75PTPPVDD306 pKa = 4.38PPATEE311 pKa = 4.58PNTPADD317 pKa = 3.83PSTPEE322 pKa = 4.0STPPTDD328 pKa = 3.97PPAPQPTPPAEE339 pKa = 4.28PSNPEE344 pKa = 3.56PTPPVDD350 pKa = 4.83PPATPPNIPADD361 pKa = 3.79PSTPEE366 pKa = 3.94STPPADD372 pKa = 4.08PPAPQPTPPAEE383 pKa = 4.4PSTPEE388 pKa = 3.56PSTPAKK394 pKa = 10.49APAPEE399 pKa = 4.31PTPPPSGPSMTPEE412 pKa = 3.78ATPPSTAGPGAEE424 pKa = 4.41TEE426 pKa = 4.45TPDD429 pKa = 5.12GDD431 pKa = 4.23TTTQPASPQTTAPMHH446 pKa = 5.9PVPDD450 pKa = 3.58ISTLLWIRR458 pKa = 11.84PTIAIILIFLLMTIFHH474 pKa = 7.05IMYY477 pKa = 8.83CVCLHH482 pKa = 6.12EE483 pKa = 4.44
MM1 pKa = 7.58CGVKK5 pKa = 10.29SLAKK9 pKa = 10.34CFLLFQIISFLGNHH23 pKa = 5.98NLVWVPGAALGAAEE37 pKa = 4.32TVEE40 pKa = 5.03GITSRR45 pKa = 11.84EE46 pKa = 3.74MEE48 pKa = 4.23INATKK53 pKa = 10.49APSSGATFSLLVTLSNNNPTTIMRR77 pKa = 11.84PPVAQNGEE85 pKa = 4.54SVHH88 pKa = 6.6KK89 pKa = 10.11DD90 pKa = 2.98ARR92 pKa = 11.84SASASDD98 pKa = 3.6PTTSEE103 pKa = 3.72PTSPGEE109 pKa = 4.34EE110 pKa = 4.11PTEE113 pKa = 4.42ADD115 pKa = 3.46PKK117 pKa = 9.53AAPSAGHH124 pKa = 5.74VGEE127 pKa = 4.99TEE129 pKa = 4.24PEE131 pKa = 4.31SPTPLPATPKK141 pKa = 10.28PSSQEE146 pKa = 4.07DD147 pKa = 4.33NPTMTPPTAEE157 pKa = 4.61PPTSNADD164 pKa = 3.03VSTEE168 pKa = 3.93HH169 pKa = 7.51VDD171 pKa = 3.35EE172 pKa = 5.04TEE174 pKa = 4.32PEE176 pKa = 4.4SPTFLPPTPEE186 pKa = 4.94PDD188 pKa = 3.38TPTTPEE194 pKa = 4.0TTTPSQNQEE203 pKa = 4.18DD204 pKa = 5.16EE205 pKa = 4.49PTLTTSSSDD214 pKa = 3.52APADD218 pKa = 3.79TSDD221 pKa = 3.37TSPPKK226 pKa = 10.48QEE228 pKa = 4.89DD229 pKa = 4.1DD230 pKa = 3.6PVKK233 pKa = 9.38PTEE236 pKa = 4.54SKK238 pKa = 10.39PQAEE242 pKa = 4.78PKK244 pKa = 10.41DD245 pKa = 3.93NSPSDD250 pKa = 3.81VPEE253 pKa = 4.39TADD256 pKa = 4.38SPTDD260 pKa = 3.51PASPTVEE267 pKa = 4.05LTPPTEE273 pKa = 4.31PPTPEE278 pKa = 4.09TVSPADD284 pKa = 3.54SPVPQPTAPAEE295 pKa = 4.03PSKK298 pKa = 10.77PEE300 pKa = 3.75PTPPVDD306 pKa = 4.38PPATEE311 pKa = 4.58PNTPADD317 pKa = 3.83PSTPEE322 pKa = 4.0STPPTDD328 pKa = 3.97PPAPQPTPPAEE339 pKa = 4.28PSNPEE344 pKa = 3.56PTPPVDD350 pKa = 4.83PPATPPNIPADD361 pKa = 3.79PSTPEE366 pKa = 3.94STPPADD372 pKa = 4.08PPAPQPTPPAEE383 pKa = 4.4PSTPEE388 pKa = 3.56PSTPAKK394 pKa = 10.49APAPEE399 pKa = 4.31PTPPPSGPSMTPEE412 pKa = 3.78ATPPSTAGPGAEE424 pKa = 4.41TEE426 pKa = 4.45TPDD429 pKa = 5.12GDD431 pKa = 4.23TTTQPASPQTTAPMHH446 pKa = 5.9PVPDD450 pKa = 3.58ISTLLWIRR458 pKa = 11.84PTIAIILIFLLMTIFHH474 pKa = 7.05IMYY477 pKa = 8.83CVCLHH482 pKa = 6.12EE483 pKa = 4.44
Molecular weight: 49.83 kDa
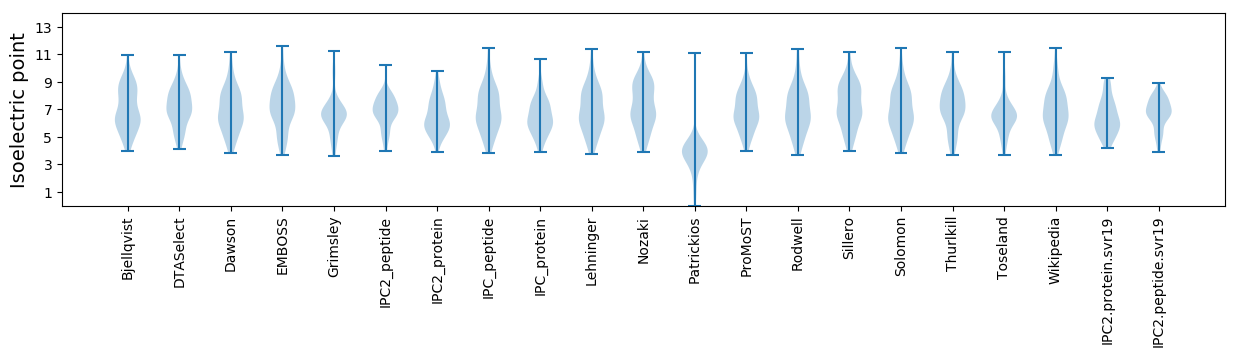
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|O41940|O41940_MHV68 Thymidine kinase OS=Murid herpesvirus 4 OX=33708 GN=GAMMAHV.ORF21 PE=3 SV=1
MM1 pKa = 7.5NEE3 pKa = 4.02LGAKK7 pKa = 9.27QLLNKK12 pKa = 9.92LPKK15 pKa = 9.66RR16 pKa = 11.84RR17 pKa = 11.84ARR19 pKa = 11.84AGKK22 pKa = 8.89LAHH25 pKa = 5.4VRR27 pKa = 11.84CYY29 pKa = 10.57RR30 pKa = 11.84AMQGASNILQLLEE43 pKa = 4.08SLKK46 pKa = 10.16IAHH49 pKa = 6.52QITHH53 pKa = 6.65PVGSRR58 pKa = 11.84LFFEE62 pKa = 4.5VTLGRR67 pKa = 11.84RR68 pKa = 11.84VVDD71 pKa = 4.63AIIVVFSEE79 pKa = 4.29SSPHH83 pKa = 4.79IHH85 pKa = 6.93CFLIEE90 pKa = 4.49LKK92 pKa = 9.19TCKK95 pKa = 10.01INFFNQHH102 pKa = 4.12STTRR106 pKa = 11.84EE107 pKa = 3.79AQKK110 pKa = 10.96VEE112 pKa = 3.84GSNQLRR118 pKa = 11.84DD119 pKa = 3.54SAKK122 pKa = 10.48ALAVLAPVGTDD133 pKa = 3.17PCRR136 pKa = 11.84VTAHH140 pKa = 7.4LIFKK144 pKa = 8.06SQRR147 pKa = 11.84GLNTLKK153 pKa = 10.68SYY155 pKa = 8.91TLNWMTHH162 pKa = 4.38TVNTQKK168 pKa = 10.73VALLNFLGLRR178 pKa = 11.84ADD180 pKa = 3.84NEE182 pKa = 3.89LRR184 pKa = 11.84ACLTRR189 pKa = 11.84GLPPANSPGSRR200 pKa = 11.84RR201 pKa = 11.84HH202 pKa = 5.56HH203 pKa = 6.06VCLPEE208 pKa = 4.44PKK210 pKa = 9.51PQKK213 pKa = 9.36HH214 pKa = 5.53LKK216 pKa = 8.59NRR218 pKa = 11.84RR219 pKa = 11.84GGAHH223 pKa = 7.14RR224 pKa = 11.84NQKK227 pKa = 9.72ARR229 pKa = 11.84RR230 pKa = 11.84QGVGPKK236 pKa = 9.95VSNDD240 pKa = 2.91KK241 pKa = 9.89TRR243 pKa = 11.84NAPTHH248 pKa = 5.79AEE250 pKa = 3.98GRR252 pKa = 11.84AGG254 pKa = 3.63
MM1 pKa = 7.5NEE3 pKa = 4.02LGAKK7 pKa = 9.27QLLNKK12 pKa = 9.92LPKK15 pKa = 9.66RR16 pKa = 11.84RR17 pKa = 11.84ARR19 pKa = 11.84AGKK22 pKa = 8.89LAHH25 pKa = 5.4VRR27 pKa = 11.84CYY29 pKa = 10.57RR30 pKa = 11.84AMQGASNILQLLEE43 pKa = 4.08SLKK46 pKa = 10.16IAHH49 pKa = 6.52QITHH53 pKa = 6.65PVGSRR58 pKa = 11.84LFFEE62 pKa = 4.5VTLGRR67 pKa = 11.84RR68 pKa = 11.84VVDD71 pKa = 4.63AIIVVFSEE79 pKa = 4.29SSPHH83 pKa = 4.79IHH85 pKa = 6.93CFLIEE90 pKa = 4.49LKK92 pKa = 9.19TCKK95 pKa = 10.01INFFNQHH102 pKa = 4.12STTRR106 pKa = 11.84EE107 pKa = 3.79AQKK110 pKa = 10.96VEE112 pKa = 3.84GSNQLRR118 pKa = 11.84DD119 pKa = 3.54SAKK122 pKa = 10.48ALAVLAPVGTDD133 pKa = 3.17PCRR136 pKa = 11.84VTAHH140 pKa = 7.4LIFKK144 pKa = 8.06SQRR147 pKa = 11.84GLNTLKK153 pKa = 10.68SYY155 pKa = 8.91TLNWMTHH162 pKa = 4.38TVNTQKK168 pKa = 10.73VALLNFLGLRR178 pKa = 11.84ADD180 pKa = 3.84NEE182 pKa = 3.89LRR184 pKa = 11.84ACLTRR189 pKa = 11.84GLPPANSPGSRR200 pKa = 11.84RR201 pKa = 11.84HH202 pKa = 5.56HH203 pKa = 6.06VCLPEE208 pKa = 4.44PKK210 pKa = 9.51PQKK213 pKa = 9.36HH214 pKa = 5.53LKK216 pKa = 8.59NRR218 pKa = 11.84RR219 pKa = 11.84GGAHH223 pKa = 7.14RR224 pKa = 11.84NQKK227 pKa = 9.72ARR229 pKa = 11.84RR230 pKa = 11.84QGVGPKK236 pKa = 9.95VSNDD240 pKa = 2.91KK241 pKa = 9.89TRR243 pKa = 11.84NAPTHH248 pKa = 5.79AEE250 pKa = 3.98GRR252 pKa = 11.84AGG254 pKa = 3.63
Molecular weight: 28.28 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
35020 |
75 |
2457 |
479.7 |
53.72 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.982 ± 0.217 | 2.613 ± 0.226 |
4.84 ± 0.131 | 5.197 ± 0.146 |
4.517 ± 0.193 | 5.086 ± 0.209 |
2.841 ± 0.102 | 6.017 ± 0.189 |
5.157 ± 0.267 | 10.528 ± 0.284 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.487 ± 0.113 | 4.417 ± 0.144 |
6.205 ± 0.401 | 3.798 ± 0.147 |
4.209 ± 0.187 | 8.198 ± 0.202 |
7.102 ± 0.24 | 6.288 ± 0.185 |
1.059 ± 0.068 | 3.458 ± 0.159 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
