
halophilic archaeon J07HX64
Taxonomy: cellular organisms; Archaea; Euryarchaeota; Stenosarchaea group; Halobacteria; Halobacteriales; unclassified Halobacteriales
Average proteome isoelectric point is 5.12
Get precalculated fractions of proteins
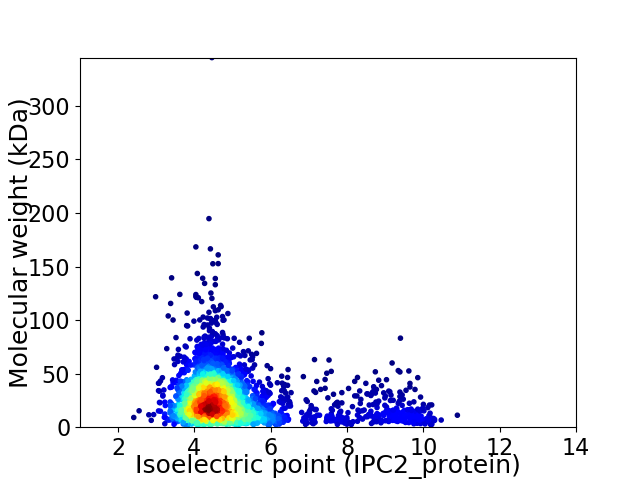
Virtual 2D-PAGE plot for 3025 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|U1QTE3|U1QTE3_9EURY Double-stranded beta-helix domain protein OS=halophilic archaeon J07HX64 OX=1085028 GN=J07HX64_02293 PE=4 SV=1
MM1 pKa = 7.44EE2 pKa = 4.86LTWHH6 pKa = 6.17GHH8 pKa = 3.98STWRR12 pKa = 11.84VEE14 pKa = 4.26LGEE17 pKa = 4.17TSMLIDD23 pKa = 4.16PFFGNPKK30 pKa = 9.25TGLSPEE36 pKa = 4.33SVEE39 pKa = 4.22TPDD42 pKa = 4.57YY43 pKa = 11.38VLLTHH48 pKa = 6.42GHH50 pKa = 6.98ADD52 pKa = 3.74HH53 pKa = 7.07IGDD56 pKa = 3.81AAAFSDD62 pKa = 3.92ATLVAVPEE70 pKa = 4.19LAAFAEE76 pKa = 4.62DD77 pKa = 3.89EE78 pKa = 4.76LGFDD82 pKa = 4.48DD83 pKa = 5.82AVAGMGMNMGGTVACGDD100 pKa = 3.92AFVTMHH106 pKa = 7.22RR107 pKa = 11.84ADD109 pKa = 3.63HH110 pKa = 6.23TNGANTGYY118 pKa = 10.4DD119 pKa = 3.9LEE121 pKa = 6.4LGMPVGYY128 pKa = 10.15VISDD132 pKa = 3.94TAPTEE137 pKa = 4.43LDD139 pKa = 3.9DD140 pKa = 5.41ADD142 pKa = 3.8STTFYY147 pKa = 10.67HH148 pKa = 7.25AGDD151 pKa = 3.73TALMTEE157 pKa = 3.86MRR159 pKa = 11.84EE160 pKa = 4.21VIGAYY165 pKa = 10.19LDD167 pKa = 3.86PDD169 pKa = 3.94AAALPAGDD177 pKa = 4.83HH178 pKa = 5.59FTMGPRR184 pKa = 11.84QAAIAAGWLGVDD196 pKa = 3.83YY197 pKa = 10.28ALPMHH202 pKa = 7.05YY203 pKa = 8.78DD204 pKa = 3.75TFPPIEE210 pKa = 4.9IDD212 pKa = 3.06TATFEE217 pKa = 5.02DD218 pKa = 4.7EE219 pKa = 4.8LGAHH223 pKa = 6.6APDD226 pKa = 3.83TEE228 pKa = 4.17AVVLDD233 pKa = 3.75GDD235 pKa = 4.11EE236 pKa = 4.41SFEE239 pKa = 4.27LL240 pKa = 4.16
MM1 pKa = 7.44EE2 pKa = 4.86LTWHH6 pKa = 6.17GHH8 pKa = 3.98STWRR12 pKa = 11.84VEE14 pKa = 4.26LGEE17 pKa = 4.17TSMLIDD23 pKa = 4.16PFFGNPKK30 pKa = 9.25TGLSPEE36 pKa = 4.33SVEE39 pKa = 4.22TPDD42 pKa = 4.57YY43 pKa = 11.38VLLTHH48 pKa = 6.42GHH50 pKa = 6.98ADD52 pKa = 3.74HH53 pKa = 7.07IGDD56 pKa = 3.81AAAFSDD62 pKa = 3.92ATLVAVPEE70 pKa = 4.19LAAFAEE76 pKa = 4.62DD77 pKa = 3.89EE78 pKa = 4.76LGFDD82 pKa = 4.48DD83 pKa = 5.82AVAGMGMNMGGTVACGDD100 pKa = 3.92AFVTMHH106 pKa = 7.22RR107 pKa = 11.84ADD109 pKa = 3.63HH110 pKa = 6.23TNGANTGYY118 pKa = 10.4DD119 pKa = 3.9LEE121 pKa = 6.4LGMPVGYY128 pKa = 10.15VISDD132 pKa = 3.94TAPTEE137 pKa = 4.43LDD139 pKa = 3.9DD140 pKa = 5.41ADD142 pKa = 3.8STTFYY147 pKa = 10.67HH148 pKa = 7.25AGDD151 pKa = 3.73TALMTEE157 pKa = 3.86MRR159 pKa = 11.84EE160 pKa = 4.21VIGAYY165 pKa = 10.19LDD167 pKa = 3.86PDD169 pKa = 3.94AAALPAGDD177 pKa = 4.83HH178 pKa = 5.59FTMGPRR184 pKa = 11.84QAAIAAGWLGVDD196 pKa = 3.83YY197 pKa = 10.28ALPMHH202 pKa = 7.05YY203 pKa = 8.78DD204 pKa = 3.75TFPPIEE210 pKa = 4.9IDD212 pKa = 3.06TATFEE217 pKa = 5.02DD218 pKa = 4.7EE219 pKa = 4.8LGAHH223 pKa = 6.6APDD226 pKa = 3.83TEE228 pKa = 4.17AVVLDD233 pKa = 3.75GDD235 pKa = 4.11EE236 pKa = 4.41SFEE239 pKa = 4.27LL240 pKa = 4.16
Molecular weight: 25.49 kDa
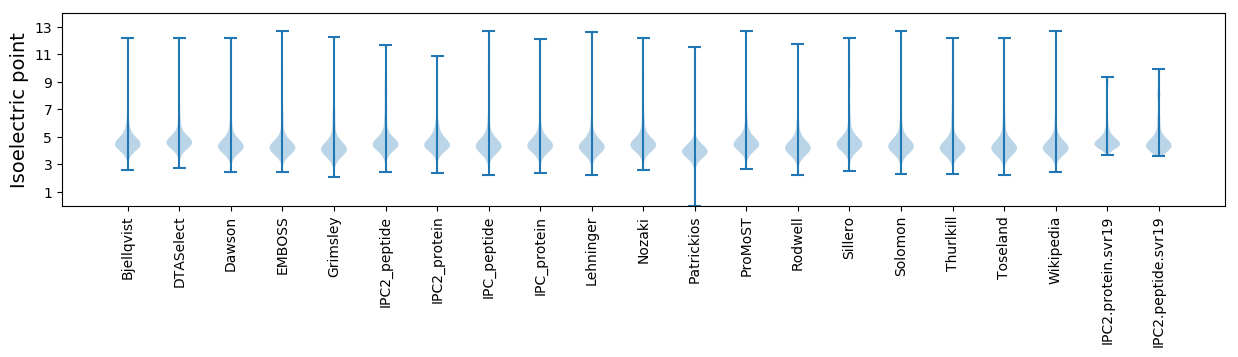
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|U1QYS5|U1QYS5_9EURY Uncharacterized protein OS=halophilic archaeon J07HX64 OX=1085028 GN=J07HX64_02918 PE=4 SV=1
MM1 pKa = 7.59CSIASTRR8 pKa = 11.84EE9 pKa = 3.12RR10 pKa = 11.84SFRR13 pKa = 11.84AKK15 pKa = 10.14RR16 pKa = 11.84GKK18 pKa = 10.25LLEE21 pKa = 4.99LGTLYY26 pKa = 11.09LDD28 pKa = 3.44VPYY31 pKa = 11.07GFVSDD36 pKa = 4.55IEE38 pKa = 5.13DD39 pKa = 4.0GEE41 pKa = 4.23QLITSSVGNHH51 pKa = 7.03DD52 pKa = 4.3LLQPASRR59 pKa = 11.84ARR61 pKa = 11.84SGSRR65 pKa = 11.84TAGRR69 pKa = 11.84RR70 pKa = 11.84SPPTTASSPSKK81 pKa = 9.9TPVEE85 pKa = 3.99AAGQRR90 pKa = 11.84TQPTRR95 pKa = 11.84CSNWRR100 pKa = 11.84RR101 pKa = 11.84TSAAGG106 pKa = 3.2
MM1 pKa = 7.59CSIASTRR8 pKa = 11.84EE9 pKa = 3.12RR10 pKa = 11.84SFRR13 pKa = 11.84AKK15 pKa = 10.14RR16 pKa = 11.84GKK18 pKa = 10.25LLEE21 pKa = 4.99LGTLYY26 pKa = 11.09LDD28 pKa = 3.44VPYY31 pKa = 11.07GFVSDD36 pKa = 4.55IEE38 pKa = 5.13DD39 pKa = 4.0GEE41 pKa = 4.23QLITSSVGNHH51 pKa = 7.03DD52 pKa = 4.3LLQPASRR59 pKa = 11.84ARR61 pKa = 11.84SGSRR65 pKa = 11.84TAGRR69 pKa = 11.84RR70 pKa = 11.84SPPTTASSPSKK81 pKa = 9.9TPVEE85 pKa = 3.99AAGQRR90 pKa = 11.84TQPTRR95 pKa = 11.84CSNWRR100 pKa = 11.84RR101 pKa = 11.84TSAAGG106 pKa = 3.2
Molecular weight: 11.42 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
767293 |
23 |
3137 |
253.7 |
27.18 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.974 ± 0.062 | 0.804 ± 0.018 |
7.865 ± 0.053 | 8.343 ± 0.068 |
3.05 ± 0.029 | 9.032 ± 0.057 |
1.993 ± 0.026 | 3.513 ± 0.034 |
1.397 ± 0.025 | 8.752 ± 0.059 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.622 ± 0.021 | 2.344 ± 0.03 |
4.777 ± 0.031 | 2.874 ± 0.029 |
7.165 ± 0.057 | 5.799 ± 0.042 |
7.062 ± 0.045 | 9.095 ± 0.059 |
1.005 ± 0.015 | 2.384 ± 0.025 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
