
Opitutae bacterium SCGC AG-212-L18
Taxonomy: cellular organisms; Bacteria; PVC group; Verrucomicrobia; Opitutae; unclassified Opitutae
Average proteome isoelectric point is 6.66
Get precalculated fractions of proteins
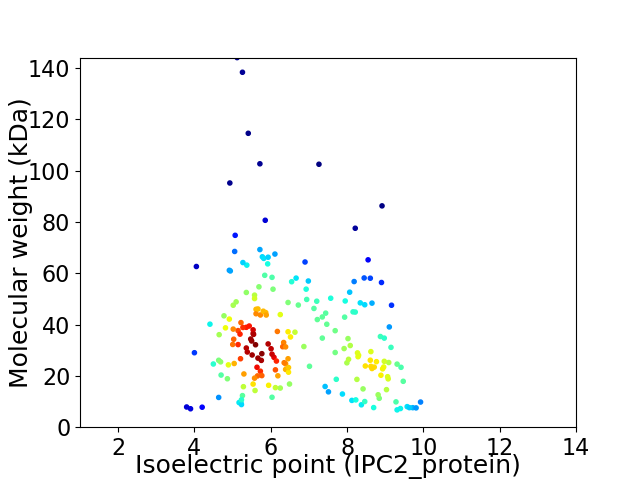
Virtual 2D-PAGE plot for 203 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A177QVD0|A0A177QVD0_9BACT Uncharacterized protein OS=Opitutae bacterium SCGC AG-212-L18 OX=1799660 GN=AYO37_00015 PE=4 SV=1
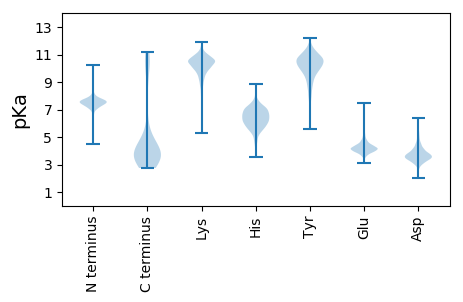
MM1 pKa = 7.61KK2 pKa = 10.15KK3 pKa = 9.81QRR5 pKa = 11.84LTIGLLIIFLLGQIDD20 pKa = 3.77VWGQRR25 pKa = 11.84HH26 pKa = 5.68EE27 pKa = 4.9GKK29 pKa = 10.39FPPVFKK35 pKa = 11.2LEE37 pKa = 5.27DD38 pKa = 3.38INGTNGYY45 pKa = 9.32IIQGLHH51 pKa = 7.13DD52 pKa = 4.87FSQQINGKK60 pKa = 10.18GDD62 pKa = 3.38VNGDD66 pKa = 3.49GVKK69 pKa = 10.54DD70 pKa = 3.66VLIGNLFLEE79 pKa = 6.06GYY81 pKa = 7.16SQRR84 pKa = 11.84DD85 pKa = 2.96QVYY88 pKa = 9.68IIFGNKK94 pKa = 7.17EE95 pKa = 3.61KK96 pKa = 10.57RR97 pKa = 11.84PQVVDD102 pKa = 3.76PLHH105 pKa = 6.69INSSNGVIVNPGMLDD120 pKa = 3.18SFGLYY125 pKa = 10.1VSVIGDD131 pKa = 3.56INKK134 pKa = 10.51DD135 pKa = 3.64GIDD138 pKa = 4.37DD139 pKa = 3.73MLISNEE145 pKa = 3.96YY146 pKa = 9.61QDD148 pKa = 5.43NFLCVLFGRR157 pKa = 11.84KK158 pKa = 8.25HH159 pKa = 5.38WSTPIVITDD168 pKa = 4.32LMDD171 pKa = 3.85GKK173 pKa = 10.55NGFFIEE179 pKa = 4.64GDD181 pKa = 3.56SNLYY185 pKa = 7.85VTAISPAGDD194 pKa = 3.33INGDD198 pKa = 3.93GIDD201 pKa = 5.04DD202 pKa = 3.53ILMGVGGNNNIQNAGQSYY220 pKa = 8.36VVFGSKK226 pKa = 10.56GSWPAVINLKK236 pKa = 10.39DD237 pKa = 3.37INGNNGFIINGIYY250 pKa = 10.61AKK252 pKa = 10.46DD253 pKa = 3.39EE254 pKa = 4.17SGYY257 pKa = 10.01SVSGAGDD264 pKa = 3.59FNGDD268 pKa = 3.87GISDD272 pKa = 4.13VIIGAPFANNGQGQSYY288 pKa = 10.29IVFGSKK294 pKa = 10.69GPWPATIDD302 pKa = 3.77LGNLDD307 pKa = 4.35GSNGLTIYY315 pKa = 10.8GIDD318 pKa = 3.58SSDD321 pKa = 3.28LTGYY325 pKa = 9.78SVSGAGDD332 pKa = 3.33INGDD336 pKa = 3.94DD337 pKa = 4.06FDD339 pKa = 5.38DD340 pKa = 5.83VIIASNNNDD349 pKa = 2.67IYY351 pKa = 11.74VVFGNNATWPPLIDD365 pKa = 4.54LSNLNGKK372 pKa = 9.28NGFVLRR378 pKa = 11.84GLYY381 pKa = 9.93KK382 pKa = 10.47RR383 pKa = 11.84GPFSVSGAGDD393 pKa = 3.77FNADD397 pKa = 3.95NITDD401 pKa = 4.59IIIGTTDD408 pKa = 2.77AYY410 pKa = 10.03IDD412 pKa = 3.16EE413 pKa = 5.31GYY415 pKa = 10.16FRR417 pKa = 11.84RR418 pKa = 11.84QGYY421 pKa = 7.85VVFGNKK427 pKa = 9.45NMWPAEE433 pKa = 3.91INLNGLNGTGLNGTNGFIINGSFNNMHH460 pKa = 5.48VRR462 pKa = 11.84YY463 pKa = 9.29VVDD466 pKa = 5.5GIGDD470 pKa = 3.72VNGDD474 pKa = 3.79SVDD477 pKa = 4.71DD478 pKa = 3.85ILIGEE483 pKa = 4.52SYY485 pKa = 10.56GANPHH490 pKa = 6.25NGVPRR495 pKa = 11.84NGYY498 pKa = 8.76VLFGCCGEE506 pKa = 4.44VPPPPLSPIEE516 pKa = 4.18PLHH519 pKa = 6.45NSLALGLGIGFGAVGLITAGVVYY542 pKa = 8.7YY543 pKa = 9.12WWHH546 pKa = 6.13YY547 pKa = 10.01DD548 pKa = 3.57DD549 pKa = 5.3PVIGADD555 pKa = 4.97DD556 pKa = 3.61IAEE559 pKa = 4.31GPINVDD565 pKa = 3.15PEE567 pKa = 4.26KK568 pKa = 10.45TLLIPIDD575 pKa = 4.32APPSGVGWMTSSS587 pKa = 3.24
MM1 pKa = 7.61KK2 pKa = 10.15KK3 pKa = 9.81QRR5 pKa = 11.84LTIGLLIIFLLGQIDD20 pKa = 3.77VWGQRR25 pKa = 11.84HH26 pKa = 5.68EE27 pKa = 4.9GKK29 pKa = 10.39FPPVFKK35 pKa = 11.2LEE37 pKa = 5.27DD38 pKa = 3.38INGTNGYY45 pKa = 9.32IIQGLHH51 pKa = 7.13DD52 pKa = 4.87FSQQINGKK60 pKa = 10.18GDD62 pKa = 3.38VNGDD66 pKa = 3.49GVKK69 pKa = 10.54DD70 pKa = 3.66VLIGNLFLEE79 pKa = 6.06GYY81 pKa = 7.16SQRR84 pKa = 11.84DD85 pKa = 2.96QVYY88 pKa = 9.68IIFGNKK94 pKa = 7.17EE95 pKa = 3.61KK96 pKa = 10.57RR97 pKa = 11.84PQVVDD102 pKa = 3.76PLHH105 pKa = 6.69INSSNGVIVNPGMLDD120 pKa = 3.18SFGLYY125 pKa = 10.1VSVIGDD131 pKa = 3.56INKK134 pKa = 10.51DD135 pKa = 3.64GIDD138 pKa = 4.37DD139 pKa = 3.73MLISNEE145 pKa = 3.96YY146 pKa = 9.61QDD148 pKa = 5.43NFLCVLFGRR157 pKa = 11.84KK158 pKa = 8.25HH159 pKa = 5.38WSTPIVITDD168 pKa = 4.32LMDD171 pKa = 3.85GKK173 pKa = 10.55NGFFIEE179 pKa = 4.64GDD181 pKa = 3.56SNLYY185 pKa = 7.85VTAISPAGDD194 pKa = 3.33INGDD198 pKa = 3.93GIDD201 pKa = 5.04DD202 pKa = 3.53ILMGVGGNNNIQNAGQSYY220 pKa = 8.36VVFGSKK226 pKa = 10.56GSWPAVINLKK236 pKa = 10.39DD237 pKa = 3.37INGNNGFIINGIYY250 pKa = 10.61AKK252 pKa = 10.46DD253 pKa = 3.39EE254 pKa = 4.17SGYY257 pKa = 10.01SVSGAGDD264 pKa = 3.59FNGDD268 pKa = 3.87GISDD272 pKa = 4.13VIIGAPFANNGQGQSYY288 pKa = 10.29IVFGSKK294 pKa = 10.69GPWPATIDD302 pKa = 3.77LGNLDD307 pKa = 4.35GSNGLTIYY315 pKa = 10.8GIDD318 pKa = 3.58SSDD321 pKa = 3.28LTGYY325 pKa = 9.78SVSGAGDD332 pKa = 3.33INGDD336 pKa = 3.94DD337 pKa = 4.06FDD339 pKa = 5.38DD340 pKa = 5.83VIIASNNNDD349 pKa = 2.67IYY351 pKa = 11.74VVFGNNATWPPLIDD365 pKa = 4.54LSNLNGKK372 pKa = 9.28NGFVLRR378 pKa = 11.84GLYY381 pKa = 9.93KK382 pKa = 10.47RR383 pKa = 11.84GPFSVSGAGDD393 pKa = 3.77FNADD397 pKa = 3.95NITDD401 pKa = 4.59IIIGTTDD408 pKa = 2.77AYY410 pKa = 10.03IDD412 pKa = 3.16EE413 pKa = 5.31GYY415 pKa = 10.16FRR417 pKa = 11.84RR418 pKa = 11.84QGYY421 pKa = 7.85VVFGNKK427 pKa = 9.45NMWPAEE433 pKa = 3.91INLNGLNGTGLNGTNGFIINGSFNNMHH460 pKa = 5.48VRR462 pKa = 11.84YY463 pKa = 9.29VVDD466 pKa = 5.5GIGDD470 pKa = 3.72VNGDD474 pKa = 3.79SVDD477 pKa = 4.71DD478 pKa = 3.85ILIGEE483 pKa = 4.52SYY485 pKa = 10.56GANPHH490 pKa = 6.25NGVPRR495 pKa = 11.84NGYY498 pKa = 8.76VLFGCCGEE506 pKa = 4.44VPPPPLSPIEE516 pKa = 4.18PLHH519 pKa = 6.45NSLALGLGIGFGAVGLITAGVVYY542 pKa = 8.7YY543 pKa = 9.12WWHH546 pKa = 6.13YY547 pKa = 10.01DD548 pKa = 3.57DD549 pKa = 5.3PVIGADD555 pKa = 4.97DD556 pKa = 3.61IAEE559 pKa = 4.31GPINVDD565 pKa = 3.15PEE567 pKa = 4.26KK568 pKa = 10.45TLLIPIDD575 pKa = 4.32APPSGVGWMTSSS587 pKa = 3.24
Molecular weight: 62.65 kDa
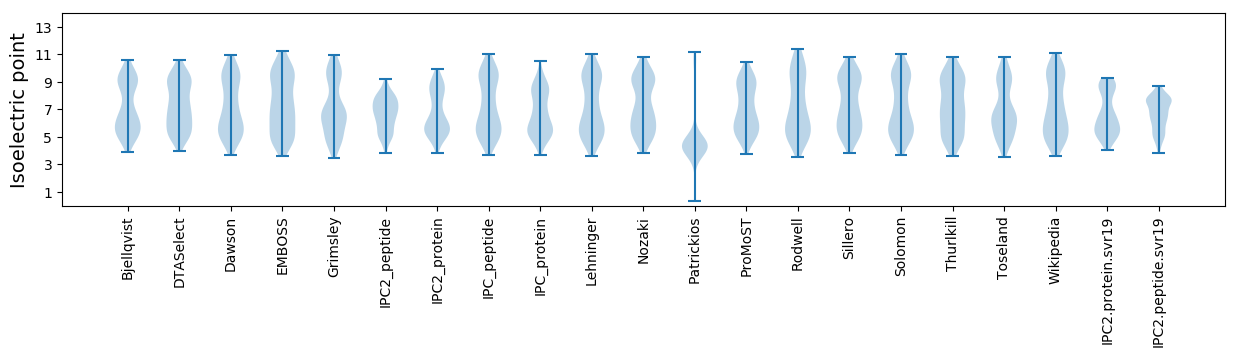
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A177QTW9|A0A177QTW9_9BACT Uncharacterized protein OS=Opitutae bacterium SCGC AG-212-L18 OX=1799660 GN=AYO37_00955 PE=4 SV=1
MM1 pKa = 7.51NIKK4 pKa = 8.76TLLVHH9 pKa = 6.39PAWLFYY15 pKa = 10.13WKK17 pKa = 9.83SQKK20 pKa = 9.89RR21 pKa = 11.84RR22 pKa = 11.84GSFLFLGRR30 pKa = 11.84MRR32 pKa = 11.84AKK34 pKa = 10.31INLRR38 pKa = 11.84LFGRR42 pKa = 11.84TFLGFDD48 pKa = 3.37EE49 pKa = 4.98YY50 pKa = 11.45GVKK53 pKa = 10.13INLRR57 pKa = 11.84KK58 pKa = 9.54EE59 pKa = 3.63WLLSYY64 pKa = 10.85NLLVIKK70 pKa = 9.0EE71 pKa = 4.12LKK73 pKa = 9.56IDD75 pKa = 3.68LVINLFF81 pKa = 3.71
MM1 pKa = 7.51NIKK4 pKa = 8.76TLLVHH9 pKa = 6.39PAWLFYY15 pKa = 10.13WKK17 pKa = 9.83SQKK20 pKa = 9.89RR21 pKa = 11.84RR22 pKa = 11.84GSFLFLGRR30 pKa = 11.84MRR32 pKa = 11.84AKK34 pKa = 10.31INLRR38 pKa = 11.84LFGRR42 pKa = 11.84TFLGFDD48 pKa = 3.37EE49 pKa = 4.98YY50 pKa = 11.45GVKK53 pKa = 10.13INLRR57 pKa = 11.84KK58 pKa = 9.54EE59 pKa = 3.63WLLSYY64 pKa = 10.85NLLVIKK70 pKa = 9.0EE71 pKa = 4.12LKK73 pKa = 9.56IDD75 pKa = 3.68LVINLFF81 pKa = 3.71
Molecular weight: 9.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
64958 |
60 |
1319 |
320.0 |
36.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.909 ± 0.263 | 1.267 ± 0.059 |
5.133 ± 0.117 | 7.219 ± 0.172 |
4.948 ± 0.129 | 5.767 ± 0.189 |
2.004 ± 0.076 | 8.164 ± 0.168 |
7.708 ± 0.189 | 10.444 ± 0.168 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.278 ± 0.082 | 5.348 ± 0.151 |
3.833 ± 0.127 | 3.687 ± 0.092 |
4.09 ± 0.128 | 6.626 ± 0.113 |
4.566 ± 0.11 | 5.427 ± 0.159 |
1.111 ± 0.061 | 3.471 ± 0.104 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
