
Musa balbisiana (Banana)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids;
Average proteome isoelectric point is 6.93
Get precalculated fractions of proteins
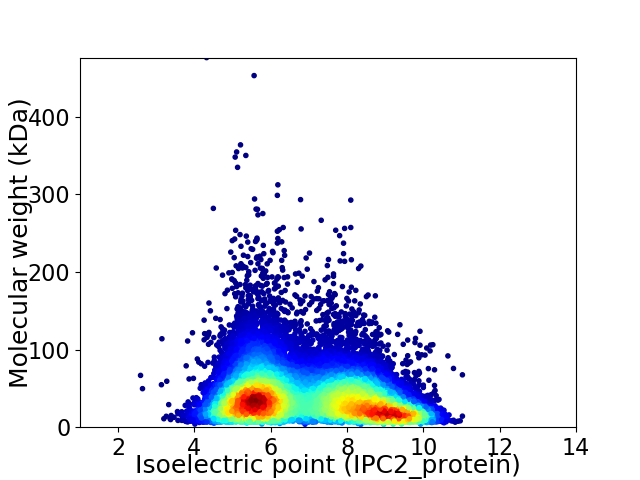
Virtual 2D-PAGE plot for 32103 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4S8IT51|A0A4S8IT51_MUSBA Uncharacterized protein ycf33 OS=Musa balbisiana OX=52838 GN=C4D60_Mb06t25590 PE=3 SV=1
MM1 pKa = 7.72LLPNYY6 pKa = 9.47CLAGSSDD13 pKa = 4.05SDD15 pKa = 3.86LAIADD20 pKa = 3.97SEE22 pKa = 4.74HH23 pKa = 6.81VVLACLDD30 pKa = 3.52LQEE33 pKa = 4.73IVEE36 pKa = 4.6SVDD39 pKa = 3.37VACNYY44 pKa = 10.36SPSQACQQQPVFDD57 pKa = 4.07VLSEE61 pKa = 4.2PEE63 pKa = 3.68IEE65 pKa = 4.73AYY67 pKa = 10.01SAEE70 pKa = 4.22PADD73 pKa = 3.9EE74 pKa = 4.46EE75 pKa = 4.59EE76 pKa = 4.32NDD78 pKa = 3.3VAYY81 pKa = 9.73MVEE84 pKa = 4.29PEE86 pKa = 4.0DD87 pKa = 5.18AIAAVMRR94 pKa = 11.84ASPAGSAGQNLNGHH108 pKa = 6.47VIAEE112 pKa = 4.2EE113 pKa = 3.97ANPAEE118 pKa = 4.36EE119 pKa = 4.11AFVWVALVAVGLALGYY135 pKa = 9.4EE136 pKa = 4.24AAVGEE141 pKa = 4.56GTVDD145 pKa = 3.66DD146 pKa = 5.09DD147 pKa = 4.08VAAEE151 pKa = 3.94AVEE154 pKa = 4.39VQTGLAIEE162 pKa = 4.36QDD164 pKa = 3.7DD165 pKa = 4.56EE166 pKa = 4.6PVVEE170 pKa = 4.99HH171 pKa = 6.49VDD173 pKa = 3.51EE174 pKa = 4.65AAVGEE179 pKa = 4.48GTVDD183 pKa = 3.66DD184 pKa = 5.09DD185 pKa = 4.16VAAEE189 pKa = 4.01AVDD192 pKa = 3.58IEE194 pKa = 4.88VPGPAAVVASEE205 pKa = 4.31DD206 pKa = 3.46KK207 pKa = 10.72TVGFDD212 pKa = 3.45VVGVADD218 pKa = 4.42QIAGLLFEE226 pKa = 5.72LGFVDD231 pKa = 6.0ALACKK236 pKa = 9.82AAEE239 pKa = 4.07QVYY242 pKa = 9.25EE243 pKa = 4.09AVVSAFAQDD252 pKa = 3.64GHH254 pKa = 5.15WVEE257 pKa = 4.71IGAGAKK263 pKa = 9.18VAAAVAVEE271 pKa = 4.34LPVIVAVAVAVAGAGLEE288 pKa = 4.09AGPGLEE294 pKa = 5.05AEE296 pKa = 4.71AEE298 pKa = 4.15LGDD301 pKa = 4.25EE302 pKa = 4.87VEE304 pKa = 4.27TAHH307 pKa = 6.11MKK309 pKa = 10.45KK310 pKa = 10.09SAAVLEE316 pKa = 4.4LLAEE320 pKa = 4.41GQLASEE326 pKa = 4.68IGHH329 pKa = 4.83TVGRR333 pKa = 11.84VAAEE337 pKa = 4.07IEE339 pKa = 4.18HH340 pKa = 6.31SVDD343 pKa = 2.65IAADD347 pKa = 3.44KK348 pKa = 10.67GHH350 pKa = 6.09SHH352 pKa = 6.02GTSDD356 pKa = 3.48STAGIAIVVAAKK368 pKa = 10.36
MM1 pKa = 7.72LLPNYY6 pKa = 9.47CLAGSSDD13 pKa = 4.05SDD15 pKa = 3.86LAIADD20 pKa = 3.97SEE22 pKa = 4.74HH23 pKa = 6.81VVLACLDD30 pKa = 3.52LQEE33 pKa = 4.73IVEE36 pKa = 4.6SVDD39 pKa = 3.37VACNYY44 pKa = 10.36SPSQACQQQPVFDD57 pKa = 4.07VLSEE61 pKa = 4.2PEE63 pKa = 3.68IEE65 pKa = 4.73AYY67 pKa = 10.01SAEE70 pKa = 4.22PADD73 pKa = 3.9EE74 pKa = 4.46EE75 pKa = 4.59EE76 pKa = 4.32NDD78 pKa = 3.3VAYY81 pKa = 9.73MVEE84 pKa = 4.29PEE86 pKa = 4.0DD87 pKa = 5.18AIAAVMRR94 pKa = 11.84ASPAGSAGQNLNGHH108 pKa = 6.47VIAEE112 pKa = 4.2EE113 pKa = 3.97ANPAEE118 pKa = 4.36EE119 pKa = 4.11AFVWVALVAVGLALGYY135 pKa = 9.4EE136 pKa = 4.24AAVGEE141 pKa = 4.56GTVDD145 pKa = 3.66DD146 pKa = 5.09DD147 pKa = 4.08VAAEE151 pKa = 3.94AVEE154 pKa = 4.39VQTGLAIEE162 pKa = 4.36QDD164 pKa = 3.7DD165 pKa = 4.56EE166 pKa = 4.6PVVEE170 pKa = 4.99HH171 pKa = 6.49VDD173 pKa = 3.51EE174 pKa = 4.65AAVGEE179 pKa = 4.48GTVDD183 pKa = 3.66DD184 pKa = 5.09DD185 pKa = 4.16VAAEE189 pKa = 4.01AVDD192 pKa = 3.58IEE194 pKa = 4.88VPGPAAVVASEE205 pKa = 4.31DD206 pKa = 3.46KK207 pKa = 10.72TVGFDD212 pKa = 3.45VVGVADD218 pKa = 4.42QIAGLLFEE226 pKa = 5.72LGFVDD231 pKa = 6.0ALACKK236 pKa = 9.82AAEE239 pKa = 4.07QVYY242 pKa = 9.25EE243 pKa = 4.09AVVSAFAQDD252 pKa = 3.64GHH254 pKa = 5.15WVEE257 pKa = 4.71IGAGAKK263 pKa = 9.18VAAAVAVEE271 pKa = 4.34LPVIVAVAVAVAGAGLEE288 pKa = 4.09AGPGLEE294 pKa = 5.05AEE296 pKa = 4.71AEE298 pKa = 4.15LGDD301 pKa = 4.25EE302 pKa = 4.87VEE304 pKa = 4.27TAHH307 pKa = 6.11MKK309 pKa = 10.45KK310 pKa = 10.09SAAVLEE316 pKa = 4.4LLAEE320 pKa = 4.41GQLASEE326 pKa = 4.68IGHH329 pKa = 4.83TVGRR333 pKa = 11.84VAAEE337 pKa = 4.07IEE339 pKa = 4.18HH340 pKa = 6.31SVDD343 pKa = 2.65IAADD347 pKa = 3.44KK348 pKa = 10.67GHH350 pKa = 6.09SHH352 pKa = 6.02GTSDD356 pKa = 3.48STAGIAIVVAAKK368 pKa = 10.36
Molecular weight: 37.37 kDa
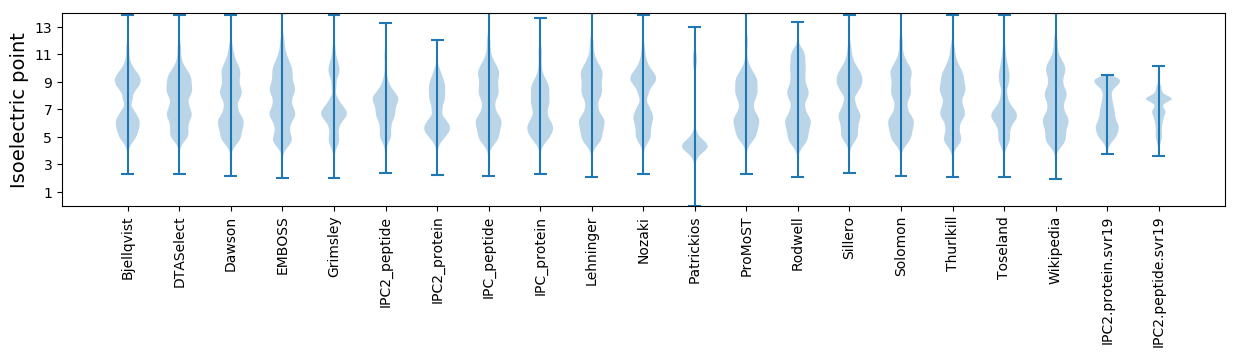
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4S8K0Q9|A0A4S8K0Q9_MUSBA Dof-type domain-containing protein OS=Musa balbisiana OX=52838 GN=C4D60_Mb08t01930 PE=4 SV=1
MM1 pKa = 7.33LVRR4 pKa = 11.84RR5 pKa = 11.84QRR7 pKa = 11.84HH8 pKa = 3.52VVRR11 pKa = 11.84RR12 pKa = 11.84GKK14 pKa = 10.29RR15 pKa = 11.84GMRR18 pKa = 11.84RR19 pKa = 11.84VLVVEE24 pKa = 4.41RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84LHH30 pKa = 6.32GIRR33 pKa = 11.84WHH35 pKa = 5.97GLGVRR40 pKa = 11.84WGKK43 pKa = 9.67RR44 pKa = 11.84GKK46 pKa = 10.42RR47 pKa = 11.84GVVVLKK53 pKa = 10.58VALRR57 pKa = 11.84VNPLLLLLRR66 pKa = 11.84LMLRR70 pKa = 11.84GNRR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84GRR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84WIVVWMVVRR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84RR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84GMGSVAAADD105 pKa = 3.53AVDD108 pKa = 4.17RR109 pKa = 11.84DD110 pKa = 3.92LHH112 pKa = 6.37HH113 pKa = 7.27LLLRR117 pKa = 4.38
MM1 pKa = 7.33LVRR4 pKa = 11.84RR5 pKa = 11.84QRR7 pKa = 11.84HH8 pKa = 3.52VVRR11 pKa = 11.84RR12 pKa = 11.84GKK14 pKa = 10.29RR15 pKa = 11.84GMRR18 pKa = 11.84RR19 pKa = 11.84VLVVEE24 pKa = 4.41RR25 pKa = 11.84RR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84LHH30 pKa = 6.32GIRR33 pKa = 11.84WHH35 pKa = 5.97GLGVRR40 pKa = 11.84WGKK43 pKa = 9.67RR44 pKa = 11.84GKK46 pKa = 10.42RR47 pKa = 11.84GVVVLKK53 pKa = 10.58VALRR57 pKa = 11.84VNPLLLLLRR66 pKa = 11.84LMLRR70 pKa = 11.84GNRR73 pKa = 11.84RR74 pKa = 11.84RR75 pKa = 11.84GRR77 pKa = 11.84RR78 pKa = 11.84RR79 pKa = 11.84RR80 pKa = 11.84WIVVWMVVRR89 pKa = 11.84RR90 pKa = 11.84RR91 pKa = 11.84RR92 pKa = 11.84RR93 pKa = 11.84RR94 pKa = 11.84RR95 pKa = 11.84RR96 pKa = 11.84GMGSVAAADD105 pKa = 3.53AVDD108 pKa = 4.17RR109 pKa = 11.84DD110 pKa = 3.92LHH112 pKa = 6.37HH113 pKa = 7.27LLLRR117 pKa = 4.38
Molecular weight: 14.23 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
12394127 |
31 |
5263 |
386.1 |
42.76 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.924 ± 0.017 | 1.908 ± 0.007 |
5.232 ± 0.009 | 6.3 ± 0.02 |
3.917 ± 0.011 | 6.958 ± 0.015 |
2.496 ± 0.006 | 4.749 ± 0.011 |
5.286 ± 0.016 | 9.715 ± 0.018 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.398 ± 0.006 | 3.687 ± 0.009 |
5.434 ± 0.017 | 3.493 ± 0.01 |
6.187 ± 0.014 | 9.103 ± 0.019 |
4.77 ± 0.008 | 6.573 ± 0.012 |
1.299 ± 0.005 | 2.572 ± 0.008 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
