
Phaeocystidibacter luteus
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Schleiferiaceae; Phaeocystidibacter
Average proteome isoelectric point is 5.76
Get precalculated fractions of proteins
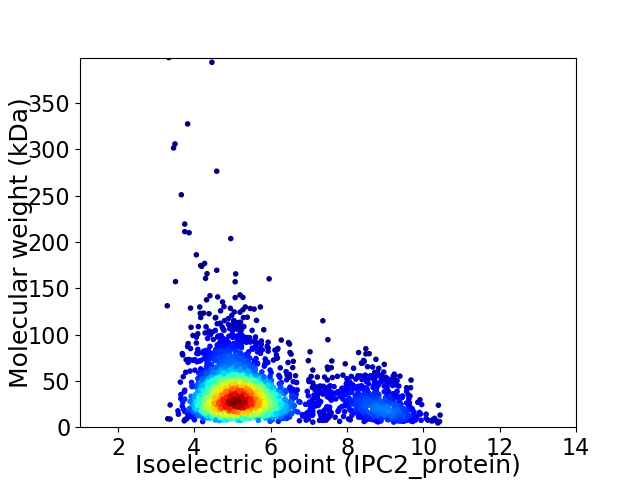
Virtual 2D-PAGE plot for 2801 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A6N6RLG2|A0A6N6RLG2_9FLAO Succinate--CoA ligase [ADP-forming] subunit beta OS=Phaeocystidibacter luteus OX=911197 GN=sucC PE=3 SV=1
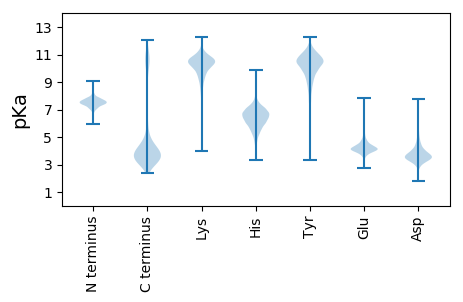
MM1 pKa = 7.71NYY3 pKa = 8.73TKK5 pKa = 10.79SFLIRR10 pKa = 11.84SMVAVAALGSFTACEE25 pKa = 4.0DD26 pKa = 3.59LSLCGLTEE34 pKa = 4.41EE35 pKa = 4.85DD36 pKa = 5.03LSMGDD41 pKa = 3.64AFVEE45 pKa = 4.51TTSYY49 pKa = 10.92FMNVVGRR56 pKa = 11.84TDD58 pKa = 2.83EE59 pKa = 4.1AMRR62 pKa = 11.84NEE64 pKa = 3.88NLQVNGSTTIDD75 pKa = 3.48GATVTRR81 pKa = 11.84TNDD84 pKa = 3.12SITIDD89 pKa = 3.55FGSNNVVTNDD99 pKa = 2.58GKK101 pKa = 8.7TRR103 pKa = 11.84RR104 pKa = 11.84GVIRR108 pKa = 11.84GYY110 pKa = 10.46ISGDD114 pKa = 3.24YY115 pKa = 9.87FQANGSINLSLEE127 pKa = 4.14NYY129 pKa = 8.74HH130 pKa = 7.03VNDD133 pKa = 3.65SLIAGSISLVNNGNNGGQDD152 pKa = 2.95PWDD155 pKa = 3.72INLSSTNFSIGNQYY169 pKa = 10.97DD170 pKa = 3.55YY171 pKa = 11.48SANLTMQWEE180 pKa = 4.3AGYY183 pKa = 8.37EE184 pKa = 4.43TIDD187 pKa = 3.33STADD191 pKa = 3.18DD192 pKa = 3.75RR193 pKa = 11.84FKK195 pKa = 11.04IFGTANGDD203 pKa = 3.93DD204 pKa = 4.4NIQLISFATSFPSPMVFDD222 pKa = 4.71RR223 pKa = 11.84SCTYY227 pKa = 10.82LVTEE231 pKa = 4.47GSVDD235 pKa = 3.38VSLTSGEE242 pKa = 4.35AEE244 pKa = 4.04VASVNVDD251 pKa = 3.87FLDD254 pKa = 4.38SDD256 pKa = 3.91GCNNVLMLNASCDD269 pKa = 3.41GTNISFPQNFDD280 pKa = 3.24GFF282 pKa = 4.04
MM1 pKa = 7.71NYY3 pKa = 8.73TKK5 pKa = 10.79SFLIRR10 pKa = 11.84SMVAVAALGSFTACEE25 pKa = 4.0DD26 pKa = 3.59LSLCGLTEE34 pKa = 4.41EE35 pKa = 4.85DD36 pKa = 5.03LSMGDD41 pKa = 3.64AFVEE45 pKa = 4.51TTSYY49 pKa = 10.92FMNVVGRR56 pKa = 11.84TDD58 pKa = 2.83EE59 pKa = 4.1AMRR62 pKa = 11.84NEE64 pKa = 3.88NLQVNGSTTIDD75 pKa = 3.48GATVTRR81 pKa = 11.84TNDD84 pKa = 3.12SITIDD89 pKa = 3.55FGSNNVVTNDD99 pKa = 2.58GKK101 pKa = 8.7TRR103 pKa = 11.84RR104 pKa = 11.84GVIRR108 pKa = 11.84GYY110 pKa = 10.46ISGDD114 pKa = 3.24YY115 pKa = 9.87FQANGSINLSLEE127 pKa = 4.14NYY129 pKa = 8.74HH130 pKa = 7.03VNDD133 pKa = 3.65SLIAGSISLVNNGNNGGQDD152 pKa = 2.95PWDD155 pKa = 3.72INLSSTNFSIGNQYY169 pKa = 10.97DD170 pKa = 3.55YY171 pKa = 11.48SANLTMQWEE180 pKa = 4.3AGYY183 pKa = 8.37EE184 pKa = 4.43TIDD187 pKa = 3.33STADD191 pKa = 3.18DD192 pKa = 3.75RR193 pKa = 11.84FKK195 pKa = 11.04IFGTANGDD203 pKa = 3.93DD204 pKa = 4.4NIQLISFATSFPSPMVFDD222 pKa = 4.71RR223 pKa = 11.84SCTYY227 pKa = 10.82LVTEE231 pKa = 4.47GSVDD235 pKa = 3.38VSLTSGEE242 pKa = 4.35AEE244 pKa = 4.04VASVNVDD251 pKa = 3.87FLDD254 pKa = 4.38SDD256 pKa = 3.91GCNNVLMLNASCDD269 pKa = 3.41GTNISFPQNFDD280 pKa = 3.24GFF282 pKa = 4.04
Molecular weight: 30.43 kDa
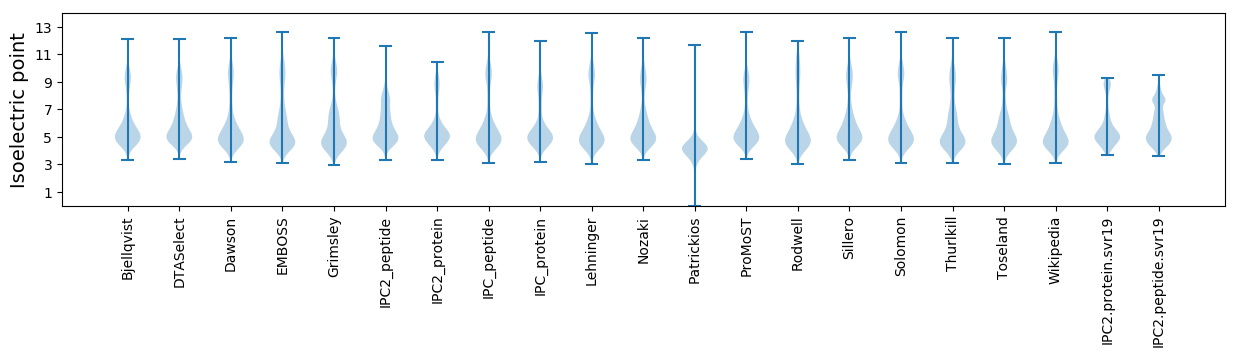
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A6N6RGI9|A0A6N6RGI9_9FLAO ATP-binding cassette domain-containing protein OS=Phaeocystidibacter luteus OX=911197 GN=F8C67_09570 PE=4 SV=1
MM1 pKa = 7.61GVRR4 pKa = 11.84KK5 pKa = 10.07LKK7 pKa = 9.13PTSPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNEE20 pKa = 3.94FSAVTTSTPEE30 pKa = 3.58KK31 pKa = 10.67SLLKK35 pKa = 10.25GKK37 pKa = 9.9KK38 pKa = 9.65RR39 pKa = 11.84SGGRR43 pKa = 11.84NNSGKK48 pKa = 8.26MTMRR52 pKa = 11.84YY53 pKa = 9.36IGGGHH58 pKa = 6.07KK59 pKa = 10.04KK60 pKa = 9.92SYY62 pKa = 10.7RR63 pKa = 11.84EE64 pKa = 3.53IDD66 pKa = 3.7FKK68 pKa = 10.97RR69 pKa = 11.84DD70 pKa = 3.08KK71 pKa = 11.42ANIPAVVKK79 pKa = 9.92TIEE82 pKa = 3.94YY83 pKa = 10.31DD84 pKa = 3.46PNRR87 pKa = 11.84TAFIALLNYY96 pKa = 9.74VDD98 pKa = 3.65GEE100 pKa = 4.02KK101 pKa = 10.32RR102 pKa = 11.84YY103 pKa = 10.22IIAPNGLQVGQEE115 pKa = 4.2VVSGSNVAPEE125 pKa = 4.7VGNSMPLSDD134 pKa = 4.54IPLGTVVSCIEE145 pKa = 3.92LRR147 pKa = 11.84PGQGAAIARR156 pKa = 11.84SAGSFAQLLARR167 pKa = 11.84DD168 pKa = 3.86GKK170 pKa = 10.29YY171 pKa = 10.63AILKK175 pKa = 8.04MPSGEE180 pKa = 3.88TRR182 pKa = 11.84MVLITCMATIGVVSNGDD199 pKa = 3.4HH200 pKa = 5.2QQVVSGKK207 pKa = 9.94AGRR210 pKa = 11.84NRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84GVAMNPVDD229 pKa = 3.68HH230 pKa = 7.13PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.4PRR245 pKa = 11.84SRR247 pKa = 11.84TGVPAKK253 pKa = 10.16GYY255 pKa = 7.56KK256 pKa = 8.87TRR258 pKa = 11.84TPKK261 pKa = 10.4KK262 pKa = 9.2ASNRR266 pKa = 11.84YY267 pKa = 8.82IIEE270 pKa = 3.97RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.17KK274 pKa = 9.82
MM1 pKa = 7.61GVRR4 pKa = 11.84KK5 pKa = 10.07LKK7 pKa = 9.13PTSPGQRR14 pKa = 11.84FRR16 pKa = 11.84VVNEE20 pKa = 3.94FSAVTTSTPEE30 pKa = 3.58KK31 pKa = 10.67SLLKK35 pKa = 10.25GKK37 pKa = 9.9KK38 pKa = 9.65RR39 pKa = 11.84SGGRR43 pKa = 11.84NNSGKK48 pKa = 8.26MTMRR52 pKa = 11.84YY53 pKa = 9.36IGGGHH58 pKa = 6.07KK59 pKa = 10.04KK60 pKa = 9.92SYY62 pKa = 10.7RR63 pKa = 11.84EE64 pKa = 3.53IDD66 pKa = 3.7FKK68 pKa = 10.97RR69 pKa = 11.84DD70 pKa = 3.08KK71 pKa = 11.42ANIPAVVKK79 pKa = 9.92TIEE82 pKa = 3.94YY83 pKa = 10.31DD84 pKa = 3.46PNRR87 pKa = 11.84TAFIALLNYY96 pKa = 9.74VDD98 pKa = 3.65GEE100 pKa = 4.02KK101 pKa = 10.32RR102 pKa = 11.84YY103 pKa = 10.22IIAPNGLQVGQEE115 pKa = 4.2VVSGSNVAPEE125 pKa = 4.7VGNSMPLSDD134 pKa = 4.54IPLGTVVSCIEE145 pKa = 3.92LRR147 pKa = 11.84PGQGAAIARR156 pKa = 11.84SAGSFAQLLARR167 pKa = 11.84DD168 pKa = 3.86GKK170 pKa = 10.29YY171 pKa = 10.63AILKK175 pKa = 8.04MPSGEE180 pKa = 3.88TRR182 pKa = 11.84MVLITCMATIGVVSNGDD199 pKa = 3.4HH200 pKa = 5.2QQVVSGKK207 pKa = 9.94AGRR210 pKa = 11.84NRR212 pKa = 11.84WLGRR216 pKa = 11.84RR217 pKa = 11.84PRR219 pKa = 11.84TRR221 pKa = 11.84GVAMNPVDD229 pKa = 3.68HH230 pKa = 7.13PMGGGEE236 pKa = 3.89GRR238 pKa = 11.84ASGGHH243 pKa = 5.4PRR245 pKa = 11.84SRR247 pKa = 11.84TGVPAKK253 pKa = 10.16GYY255 pKa = 7.56KK256 pKa = 8.87TRR258 pKa = 11.84TPKK261 pKa = 10.4KK262 pKa = 9.2ASNRR266 pKa = 11.84YY267 pKa = 8.82IIEE270 pKa = 3.97RR271 pKa = 11.84RR272 pKa = 11.84KK273 pKa = 10.17KK274 pKa = 9.82
Molecular weight: 29.67 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
972285 |
38 |
3987 |
347.1 |
38.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.283 ± 0.055 | 0.722 ± 0.017 |
5.933 ± 0.032 | 6.889 ± 0.06 |
4.772 ± 0.038 | 7.304 ± 0.053 |
2.013 ± 0.023 | 6.542 ± 0.038 |
4.931 ± 0.063 | 9.095 ± 0.061 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.518 ± 0.026 | 4.856 ± 0.041 |
3.91 ± 0.031 | 3.428 ± 0.026 |
4.772 ± 0.044 | 6.983 ± 0.056 |
5.79 ± 0.073 | 6.953 ± 0.039 |
1.419 ± 0.022 | 3.888 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
