
Tuhoko virus 2
Taxonomy: Viruses; Riboviria; Orthornavirae; Negarnaviricota; Haploviricotina; Monjiviricetes; Mononegavirales; Paramyxoviridae; Rubulavirinae; Pararubulavirus; Tuhoko pararubulavirus 2
Average proteome isoelectric point is 6.71
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
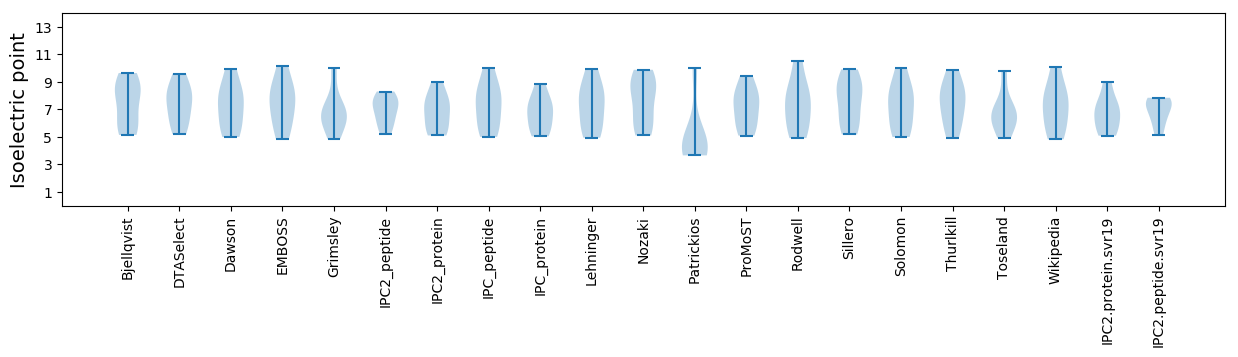
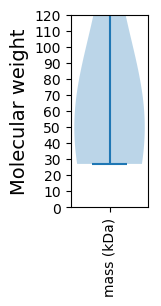
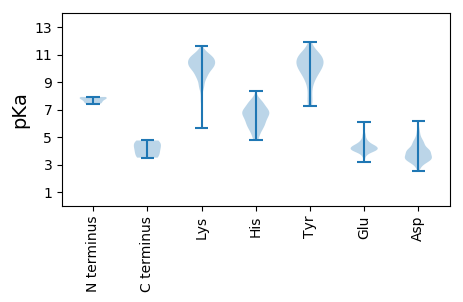
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D8WJ30|D8WJ30_9MONO Phosphoprotein OS=Tuhoko virus 2 OX=798073 GN=V/P PE=3 SV=1
MM1 pKa = 7.48SSVFQAFEE9 pKa = 4.07QFTLEE14 pKa = 3.65QDD16 pKa = 3.61QQVGGGNLEE25 pKa = 4.19VPPEE29 pKa = 4.04TLQTTIKK36 pKa = 10.68VFILNTQNPQIRR48 pKa = 11.84YY49 pKa = 9.52QMLCFCLRR57 pKa = 11.84IIASNSARR65 pKa = 11.84TAQKK69 pKa = 10.34HH70 pKa = 4.8GALLTILSLPTAMMQNHH87 pKa = 5.88IRR89 pKa = 11.84IADD92 pKa = 4.23RR93 pKa = 11.84SPDD96 pKa = 3.55SVIEE100 pKa = 4.4RR101 pKa = 11.84IEE103 pKa = 3.87IEE105 pKa = 4.1GFEE108 pKa = 4.41PGTYY112 pKa = 9.72RR113 pKa = 11.84LRR115 pKa = 11.84ANARR119 pKa = 11.84TPMTAGEE126 pKa = 4.39VIALEE131 pKa = 4.13NMAEE135 pKa = 4.24DD136 pKa = 5.5LPDD139 pKa = 3.44TLANQTPFLNPNTEE153 pKa = 4.97DD154 pKa = 3.96EE155 pKa = 4.67ICDD158 pKa = 3.68EE159 pKa = 4.22MEE161 pKa = 4.09KK162 pKa = 10.78FLDD165 pKa = 3.45AVYY168 pKa = 10.83SVLIQVWVMVCKK180 pKa = 10.77AMTAFDD186 pKa = 4.13QPTGSDD192 pKa = 2.96TRR194 pKa = 11.84RR195 pKa = 11.84LAKK198 pKa = 10.1YY199 pKa = 7.72QQQGRR204 pKa = 11.84MDD206 pKa = 3.61QKK208 pKa = 11.24YY209 pKa = 10.1LLQNEE214 pKa = 4.29VRR216 pKa = 11.84RR217 pKa = 11.84LIQLGIRR224 pKa = 11.84EE225 pKa = 4.55SLTIRR230 pKa = 11.84QFLVFEE236 pKa = 4.25MQTARR241 pKa = 11.84RR242 pKa = 11.84QGPITSKK249 pKa = 9.45YY250 pKa = 8.55YY251 pKa = 11.74AMIGDD256 pKa = 3.91IARR259 pKa = 11.84YY260 pKa = 9.21IEE262 pKa = 4.13NAGMGPFFMTIKK274 pKa = 10.32FALGTRR280 pKa = 11.84WPPLALAAFSGEE292 pKa = 4.01LVKK295 pKa = 10.85IKK297 pKa = 10.93GLMQLYY303 pKa = 9.93RR304 pKa = 11.84RR305 pKa = 11.84LGDD308 pKa = 3.29RR309 pKa = 11.84AKK311 pKa = 11.15YY312 pKa = 8.54MALLEE317 pKa = 4.21MSEE320 pKa = 4.12XMDD323 pKa = 4.67FAPANYY329 pKa = 9.71QLLFSYY335 pKa = 11.03AMGIGRR341 pKa = 11.84VQDD344 pKa = 3.84PQLQGYY350 pKa = 8.67NFARR354 pKa = 11.84PFLNAAFFQLGMEE367 pKa = 4.37TANRR371 pKa = 11.84QQGSVDD377 pKa = 3.22RR378 pKa = 11.84EE379 pKa = 3.97MAEE382 pKa = 4.08EE383 pKa = 4.8LGLTDD388 pKa = 3.67ADD390 pKa = 3.89KK391 pKa = 11.59RR392 pKa = 11.84EE393 pKa = 4.09MAAQLTRR400 pKa = 11.84LTTGRR405 pKa = 11.84GGDD408 pKa = 3.71DD409 pKa = 2.95QQGIVNVFGRR419 pKa = 11.84RR420 pKa = 11.84AQRR423 pKa = 11.84QQVIQNQDD431 pKa = 2.63QFRR434 pKa = 11.84VVEE437 pKa = 4.16EE438 pKa = 4.31EE439 pKa = 3.8EE440 pKa = 4.36SEE442 pKa = 4.23EE443 pKa = 3.95EE444 pKa = 3.95EE445 pKa = 4.79EE446 pKa = 4.53EE447 pKa = 4.77IEE449 pKa = 4.14EE450 pKa = 4.18QPVRR454 pKa = 11.84GAAPAPIRR462 pKa = 11.84RR463 pKa = 11.84PNDD466 pKa = 3.07EE467 pKa = 3.64EE468 pKa = 4.45AAWQARR474 pKa = 11.84LNEE477 pKa = 4.12LEE479 pKa = 3.99GMQRR483 pKa = 11.84RR484 pKa = 11.84PHH486 pKa = 6.7PPRR489 pKa = 11.84HH490 pKa = 5.34VQLHH494 pKa = 6.23PLQPVPVGLAAAQIHH509 pKa = 6.29RR510 pKa = 11.84EE511 pKa = 3.98PPAQEE516 pKa = 4.05LVADD520 pKa = 4.47LVEE523 pKa = 4.12
MM1 pKa = 7.48SSVFQAFEE9 pKa = 4.07QFTLEE14 pKa = 3.65QDD16 pKa = 3.61QQVGGGNLEE25 pKa = 4.19VPPEE29 pKa = 4.04TLQTTIKK36 pKa = 10.68VFILNTQNPQIRR48 pKa = 11.84YY49 pKa = 9.52QMLCFCLRR57 pKa = 11.84IIASNSARR65 pKa = 11.84TAQKK69 pKa = 10.34HH70 pKa = 4.8GALLTILSLPTAMMQNHH87 pKa = 5.88IRR89 pKa = 11.84IADD92 pKa = 4.23RR93 pKa = 11.84SPDD96 pKa = 3.55SVIEE100 pKa = 4.4RR101 pKa = 11.84IEE103 pKa = 3.87IEE105 pKa = 4.1GFEE108 pKa = 4.41PGTYY112 pKa = 9.72RR113 pKa = 11.84LRR115 pKa = 11.84ANARR119 pKa = 11.84TPMTAGEE126 pKa = 4.39VIALEE131 pKa = 4.13NMAEE135 pKa = 4.24DD136 pKa = 5.5LPDD139 pKa = 3.44TLANQTPFLNPNTEE153 pKa = 4.97DD154 pKa = 3.96EE155 pKa = 4.67ICDD158 pKa = 3.68EE159 pKa = 4.22MEE161 pKa = 4.09KK162 pKa = 10.78FLDD165 pKa = 3.45AVYY168 pKa = 10.83SVLIQVWVMVCKK180 pKa = 10.77AMTAFDD186 pKa = 4.13QPTGSDD192 pKa = 2.96TRR194 pKa = 11.84RR195 pKa = 11.84LAKK198 pKa = 10.1YY199 pKa = 7.72QQQGRR204 pKa = 11.84MDD206 pKa = 3.61QKK208 pKa = 11.24YY209 pKa = 10.1LLQNEE214 pKa = 4.29VRR216 pKa = 11.84RR217 pKa = 11.84LIQLGIRR224 pKa = 11.84EE225 pKa = 4.55SLTIRR230 pKa = 11.84QFLVFEE236 pKa = 4.25MQTARR241 pKa = 11.84RR242 pKa = 11.84QGPITSKK249 pKa = 9.45YY250 pKa = 8.55YY251 pKa = 11.74AMIGDD256 pKa = 3.91IARR259 pKa = 11.84YY260 pKa = 9.21IEE262 pKa = 4.13NAGMGPFFMTIKK274 pKa = 10.32FALGTRR280 pKa = 11.84WPPLALAAFSGEE292 pKa = 4.01LVKK295 pKa = 10.85IKK297 pKa = 10.93GLMQLYY303 pKa = 9.93RR304 pKa = 11.84RR305 pKa = 11.84LGDD308 pKa = 3.29RR309 pKa = 11.84AKK311 pKa = 11.15YY312 pKa = 8.54MALLEE317 pKa = 4.21MSEE320 pKa = 4.12XMDD323 pKa = 4.67FAPANYY329 pKa = 9.71QLLFSYY335 pKa = 11.03AMGIGRR341 pKa = 11.84VQDD344 pKa = 3.84PQLQGYY350 pKa = 8.67NFARR354 pKa = 11.84PFLNAAFFQLGMEE367 pKa = 4.37TANRR371 pKa = 11.84QQGSVDD377 pKa = 3.22RR378 pKa = 11.84EE379 pKa = 3.97MAEE382 pKa = 4.08EE383 pKa = 4.8LGLTDD388 pKa = 3.67ADD390 pKa = 3.89KK391 pKa = 11.59RR392 pKa = 11.84EE393 pKa = 4.09MAAQLTRR400 pKa = 11.84LTTGRR405 pKa = 11.84GGDD408 pKa = 3.71DD409 pKa = 2.95QQGIVNVFGRR419 pKa = 11.84RR420 pKa = 11.84AQRR423 pKa = 11.84QQVIQNQDD431 pKa = 2.63QFRR434 pKa = 11.84VVEE437 pKa = 4.16EE438 pKa = 4.31EE439 pKa = 3.8EE440 pKa = 4.36SEE442 pKa = 4.23EE443 pKa = 3.95EE444 pKa = 3.95EE445 pKa = 4.79EE446 pKa = 4.53EE447 pKa = 4.77IEE449 pKa = 4.14EE450 pKa = 4.18QPVRR454 pKa = 11.84GAAPAPIRR462 pKa = 11.84RR463 pKa = 11.84PNDD466 pKa = 3.07EE467 pKa = 3.64EE468 pKa = 4.45AAWQARR474 pKa = 11.84LNEE477 pKa = 4.12LEE479 pKa = 3.99GMQRR483 pKa = 11.84RR484 pKa = 11.84PHH486 pKa = 6.7PPRR489 pKa = 11.84HH490 pKa = 5.34VQLHH494 pKa = 6.23PLQPVPVGLAAAQIHH509 pKa = 6.29RR510 pKa = 11.84EE511 pKa = 3.98PPAQEE516 pKa = 4.05LVADD520 pKa = 4.47LVEE523 pKa = 4.12
Molecular weight: 59.24 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D8WJ33|D8WJ33_9MONO Fusion glycoprotein F0 OS=Tuhoko virus 2 OX=798073 GN=F PE=3 SV=1
MM1 pKa = 7.76AHH3 pKa = 6.39RR4 pKa = 11.84QATVSIKK11 pKa = 10.54VNHH14 pKa = 6.07EE15 pKa = 3.85SEE17 pKa = 4.53KK18 pKa = 9.89NHH20 pKa = 6.12LRR22 pKa = 11.84PFPIVQLPPSPGGSRR37 pKa = 11.84GRR39 pKa = 11.84LTRR42 pKa = 11.84QIRR45 pKa = 11.84IKK47 pKa = 10.74DD48 pKa = 3.6LTPAGSTEE56 pKa = 3.91PSLTFINTYY65 pKa = 10.58GFAKK69 pKa = 10.18PLRR72 pKa = 11.84TRR74 pKa = 11.84TEE76 pKa = 4.15FFSEE80 pKa = 3.77LHH82 pKa = 6.59KK83 pKa = 10.64PDD85 pKa = 3.98SLPSVTACTLPFGAGPNIEE104 pKa = 4.27HH105 pKa = 7.61PMRR108 pKa = 11.84LLDD111 pKa = 4.75EE112 pKa = 4.39IEE114 pKa = 4.14KK115 pKa = 10.19VHH117 pKa = 7.59IVMRR121 pKa = 11.84KK122 pKa = 7.68SASARR127 pKa = 11.84EE128 pKa = 3.87EE129 pKa = 3.73IVFNVCRR136 pKa = 11.84LPPILSKK143 pKa = 10.77HH144 pKa = 6.29HH145 pKa = 6.72IATQKK150 pKa = 10.19LVCISSEE157 pKa = 4.34KK158 pKa = 9.82YY159 pKa = 10.35LKK161 pKa = 10.93APGKK165 pKa = 9.23MISGMDD171 pKa = 3.79YY172 pKa = 9.66NYY174 pKa = 10.24HH175 pKa = 5.96IAFISVVYY183 pKa = 10.16CPASLKK189 pKa = 10.19FRR191 pKa = 11.84VMRR194 pKa = 11.84PLQLLRR200 pKa = 11.84SSTMRR205 pKa = 11.84SIQLEE210 pKa = 4.42VILVIEE216 pKa = 4.9CSDD219 pKa = 3.48NSPATRR225 pKa = 11.84NLIFDD230 pKa = 4.98DD231 pKa = 4.89EE232 pKa = 5.04SKK234 pKa = 8.87TWRR237 pKa = 11.84ASVWFHH243 pKa = 6.53LCNILKK249 pKa = 10.31SNKK252 pKa = 9.47SAEE255 pKa = 4.41KK256 pKa = 10.87YY257 pKa = 9.83DD258 pKa = 3.36DD259 pKa = 4.2HH260 pKa = 8.34YY261 pKa = 11.87FNEE264 pKa = 4.53KK265 pKa = 8.69CKK267 pKa = 10.89KK268 pKa = 8.52MDD270 pKa = 4.06LEE272 pKa = 4.46VGIADD277 pKa = 3.19MWGPTFLVKK286 pKa = 10.59AHH288 pKa = 5.9GKK290 pKa = 8.94IPKK293 pKa = 8.06TAQVYY298 pKa = 7.28FSPKK302 pKa = 9.38GWSCHH307 pKa = 5.53PLVDD311 pKa = 4.92AAPALSKK318 pKa = 10.7ILWSTGARR326 pKa = 11.84IVNVNAILQPSDD338 pKa = 3.52LGQLVQVSDD347 pKa = 3.81VIYY350 pKa = 9.81PKK352 pKa = 10.98VKK354 pKa = 10.12INKK357 pKa = 9.27KK358 pKa = 10.27LMNTAPSRR366 pKa = 11.84WNPVKK371 pKa = 10.6KK372 pKa = 10.45AVLAA376 pKa = 4.44
MM1 pKa = 7.76AHH3 pKa = 6.39RR4 pKa = 11.84QATVSIKK11 pKa = 10.54VNHH14 pKa = 6.07EE15 pKa = 3.85SEE17 pKa = 4.53KK18 pKa = 9.89NHH20 pKa = 6.12LRR22 pKa = 11.84PFPIVQLPPSPGGSRR37 pKa = 11.84GRR39 pKa = 11.84LTRR42 pKa = 11.84QIRR45 pKa = 11.84IKK47 pKa = 10.74DD48 pKa = 3.6LTPAGSTEE56 pKa = 3.91PSLTFINTYY65 pKa = 10.58GFAKK69 pKa = 10.18PLRR72 pKa = 11.84TRR74 pKa = 11.84TEE76 pKa = 4.15FFSEE80 pKa = 3.77LHH82 pKa = 6.59KK83 pKa = 10.64PDD85 pKa = 3.98SLPSVTACTLPFGAGPNIEE104 pKa = 4.27HH105 pKa = 7.61PMRR108 pKa = 11.84LLDD111 pKa = 4.75EE112 pKa = 4.39IEE114 pKa = 4.14KK115 pKa = 10.19VHH117 pKa = 7.59IVMRR121 pKa = 11.84KK122 pKa = 7.68SASARR127 pKa = 11.84EE128 pKa = 3.87EE129 pKa = 3.73IVFNVCRR136 pKa = 11.84LPPILSKK143 pKa = 10.77HH144 pKa = 6.29HH145 pKa = 6.72IATQKK150 pKa = 10.19LVCISSEE157 pKa = 4.34KK158 pKa = 9.82YY159 pKa = 10.35LKK161 pKa = 10.93APGKK165 pKa = 9.23MISGMDD171 pKa = 3.79YY172 pKa = 9.66NYY174 pKa = 10.24HH175 pKa = 5.96IAFISVVYY183 pKa = 10.16CPASLKK189 pKa = 10.19FRR191 pKa = 11.84VMRR194 pKa = 11.84PLQLLRR200 pKa = 11.84SSTMRR205 pKa = 11.84SIQLEE210 pKa = 4.42VILVIEE216 pKa = 4.9CSDD219 pKa = 3.48NSPATRR225 pKa = 11.84NLIFDD230 pKa = 4.98DD231 pKa = 4.89EE232 pKa = 5.04SKK234 pKa = 8.87TWRR237 pKa = 11.84ASVWFHH243 pKa = 6.53LCNILKK249 pKa = 10.31SNKK252 pKa = 9.47SAEE255 pKa = 4.41KK256 pKa = 10.87YY257 pKa = 9.83DD258 pKa = 3.36DD259 pKa = 4.2HH260 pKa = 8.34YY261 pKa = 11.87FNEE264 pKa = 4.53KK265 pKa = 8.69CKK267 pKa = 10.89KK268 pKa = 8.52MDD270 pKa = 4.06LEE272 pKa = 4.46VGIADD277 pKa = 3.19MWGPTFLVKK286 pKa = 10.59AHH288 pKa = 5.9GKK290 pKa = 8.94IPKK293 pKa = 8.06TAQVYY298 pKa = 7.28FSPKK302 pKa = 9.38GWSCHH307 pKa = 5.53PLVDD311 pKa = 4.92AAPALSKK318 pKa = 10.7ILWSTGARR326 pKa = 11.84IVNVNAILQPSDD338 pKa = 3.52LGQLVQVSDD347 pKa = 3.81VIYY350 pKa = 9.81PKK352 pKa = 10.98VKK354 pKa = 10.12INKK357 pKa = 9.27KK358 pKa = 10.27LMNTAPSRR366 pKa = 11.84WNPVKK371 pKa = 10.6KK372 pKa = 10.45AVLAA376 pKa = 4.44
Molecular weight: 42.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4935 |
241 |
2267 |
705.0 |
79.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.856 ± 0.671 | 2.006 ± 0.231 |
5.187 ± 0.462 | 5.593 ± 0.724 |
3.465 ± 0.229 | 5.046 ± 0.514 |
1.803 ± 0.214 | 8.024 ± 0.791 |
5.471 ± 0.555 | 10.902 ± 1.027 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.492 ± 0.25 | 4.985 ± 0.46 |
5.431 ± 0.707 | 5.187 ± 0.751 |
4.965 ± 0.585 | 8.065 ± 0.65 |
5.876 ± 0.303 | 5.512 ± 0.522 |
0.952 ± 0.204 | 3.161 ± 0.401 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
