
Hubei tombus-like virus 39
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.02
Get precalculated fractions of proteins
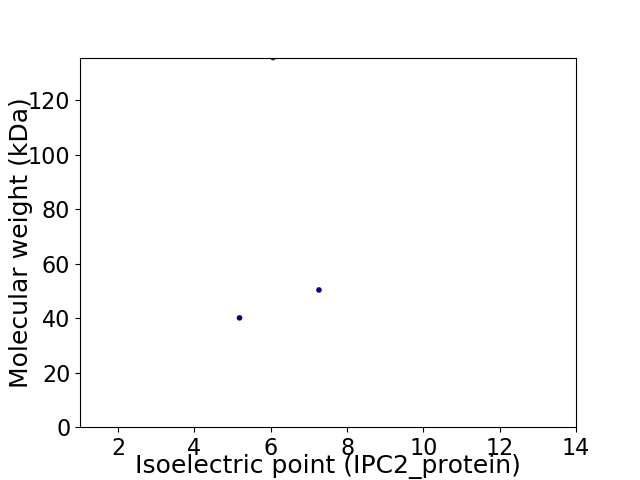
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KGI9|A0A1L3KGI9_9VIRU RNA-directed RNA polymerase OS=Hubei tombus-like virus 39 OX=1923287 PE=4 SV=1
MM1 pKa = 7.72TILKK5 pKa = 10.57GKK7 pKa = 9.16VCSINSNEE15 pKa = 4.42AVTCQLFHH23 pKa = 6.75AAAFRR28 pKa = 11.84RR29 pKa = 11.84ILMSFDD35 pKa = 3.13GFQIQSKK42 pKa = 10.45LKK44 pKa = 10.57LNYY47 pKa = 9.74EE48 pKa = 3.87PSSRR52 pKa = 11.84LNAGVVGVIVTVGIKK67 pKa = 10.23CAMEE71 pKa = 4.32NPTDD75 pKa = 3.5VCQYY79 pKa = 10.46APHH82 pKa = 6.67AAEE85 pKa = 4.71GMGGQANTSHH95 pKa = 5.96SMKK98 pKa = 10.35YY99 pKa = 10.48DD100 pKa = 4.43LAPMDD105 pKa = 4.41LVLEE109 pKa = 4.5SEE111 pKa = 4.02LHH113 pKa = 5.12EE114 pKa = 4.33RR115 pKa = 11.84MVYY118 pKa = 8.47VTVAGLDD125 pKa = 4.37SEE127 pKa = 4.99DD128 pKa = 3.51HH129 pKa = 6.01WLGDD133 pKa = 3.56LTYY136 pKa = 9.48EE137 pKa = 3.9LHH139 pKa = 5.26VQEE142 pKa = 5.65RR143 pKa = 11.84YY144 pKa = 9.12PSPIALFAVNNSIRR158 pKa = 11.84MANCKK163 pKa = 9.95RR164 pKa = 11.84SVSGTTYY171 pKa = 10.59TNVLATTTTIHH182 pKa = 6.56SSNHH186 pKa = 6.02DD187 pKa = 3.34SSGFSTGVGSSITEE201 pKa = 4.14LEE203 pKa = 3.89VDD205 pKa = 4.51SIVTNNIQTLWFNVRR220 pKa = 11.84TCGYY224 pKa = 7.0NTLYY228 pKa = 10.84FIGIGSLGLNFVLSNFNDD246 pKa = 3.61TDD248 pKa = 3.7GVEE251 pKa = 4.54VIDD254 pKa = 5.1SGEE257 pKa = 3.96IDD259 pKa = 4.98DD260 pKa = 5.91DD261 pKa = 5.32PNTSWYY267 pKa = 7.91ITFYY271 pKa = 10.68CRR273 pKa = 11.84KK274 pKa = 6.67TTDD277 pKa = 3.04IPLLSVDD284 pKa = 3.33QTTDD288 pKa = 2.96GFLVEE293 pKa = 4.66YY294 pKa = 9.55PFLLSTVTYY303 pKa = 9.76GSPVEE308 pKa = 4.77HH309 pKa = 6.2NTSVAKK315 pKa = 10.3VRR317 pKa = 11.84NLNKK321 pKa = 9.9HH322 pKa = 5.06QLAMLPPTCILPATTSHH339 pKa = 7.1SDD341 pKa = 3.08TSDD344 pKa = 2.95NRR346 pKa = 11.84PQHH349 pKa = 4.39QAFRR353 pKa = 11.84HH354 pKa = 4.89TEE356 pKa = 3.88ATKK359 pKa = 8.52TTYY362 pKa = 10.96SKK364 pKa = 11.56
MM1 pKa = 7.72TILKK5 pKa = 10.57GKK7 pKa = 9.16VCSINSNEE15 pKa = 4.42AVTCQLFHH23 pKa = 6.75AAAFRR28 pKa = 11.84RR29 pKa = 11.84ILMSFDD35 pKa = 3.13GFQIQSKK42 pKa = 10.45LKK44 pKa = 10.57LNYY47 pKa = 9.74EE48 pKa = 3.87PSSRR52 pKa = 11.84LNAGVVGVIVTVGIKK67 pKa = 10.23CAMEE71 pKa = 4.32NPTDD75 pKa = 3.5VCQYY79 pKa = 10.46APHH82 pKa = 6.67AAEE85 pKa = 4.71GMGGQANTSHH95 pKa = 5.96SMKK98 pKa = 10.35YY99 pKa = 10.48DD100 pKa = 4.43LAPMDD105 pKa = 4.41LVLEE109 pKa = 4.5SEE111 pKa = 4.02LHH113 pKa = 5.12EE114 pKa = 4.33RR115 pKa = 11.84MVYY118 pKa = 8.47VTVAGLDD125 pKa = 4.37SEE127 pKa = 4.99DD128 pKa = 3.51HH129 pKa = 6.01WLGDD133 pKa = 3.56LTYY136 pKa = 9.48EE137 pKa = 3.9LHH139 pKa = 5.26VQEE142 pKa = 5.65RR143 pKa = 11.84YY144 pKa = 9.12PSPIALFAVNNSIRR158 pKa = 11.84MANCKK163 pKa = 9.95RR164 pKa = 11.84SVSGTTYY171 pKa = 10.59TNVLATTTTIHH182 pKa = 6.56SSNHH186 pKa = 6.02DD187 pKa = 3.34SSGFSTGVGSSITEE201 pKa = 4.14LEE203 pKa = 3.89VDD205 pKa = 4.51SIVTNNIQTLWFNVRR220 pKa = 11.84TCGYY224 pKa = 7.0NTLYY228 pKa = 10.84FIGIGSLGLNFVLSNFNDD246 pKa = 3.61TDD248 pKa = 3.7GVEE251 pKa = 4.54VIDD254 pKa = 5.1SGEE257 pKa = 3.96IDD259 pKa = 4.98DD260 pKa = 5.91DD261 pKa = 5.32PNTSWYY267 pKa = 7.91ITFYY271 pKa = 10.68CRR273 pKa = 11.84KK274 pKa = 6.67TTDD277 pKa = 3.04IPLLSVDD284 pKa = 3.33QTTDD288 pKa = 2.96GFLVEE293 pKa = 4.66YY294 pKa = 9.55PFLLSTVTYY303 pKa = 9.76GSPVEE308 pKa = 4.77HH309 pKa = 6.2NTSVAKK315 pKa = 10.3VRR317 pKa = 11.84NLNKK321 pKa = 9.9HH322 pKa = 5.06QLAMLPPTCILPATTSHH339 pKa = 7.1SDD341 pKa = 3.08TSDD344 pKa = 2.95NRR346 pKa = 11.84PQHH349 pKa = 4.39QAFRR353 pKa = 11.84HH354 pKa = 4.89TEE356 pKa = 3.88ATKK359 pKa = 8.52TTYY362 pKa = 10.96SKK364 pKa = 11.56
Molecular weight: 40.1 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KGP9|A0A1L3KGP9_9VIRU Chropara_Vmeth domain-containing protein OS=Hubei tombus-like virus 39 OX=1923287 PE=4 SV=1
MM1 pKa = 7.74EE2 pKa = 5.01FPTWVARR9 pKa = 11.84YY10 pKa = 7.18TGGRR14 pKa = 11.84RR15 pKa = 11.84KK16 pKa = 9.4QLQRR20 pKa = 11.84EE21 pKa = 4.1FDD23 pKa = 3.37TGHH26 pKa = 6.44WKK28 pKa = 10.46VADD31 pKa = 3.91EE32 pKa = 4.35AASGVKK38 pKa = 10.26SFIKK42 pKa = 10.27IEE44 pKa = 4.23AGKK47 pKa = 8.27PTDD50 pKa = 3.91PRR52 pKa = 11.84NISPRR57 pKa = 11.84PDD59 pKa = 3.08EE60 pKa = 4.02YY61 pKa = 11.14LVRR64 pKa = 11.84LGPIIAGIEE73 pKa = 3.92RR74 pKa = 11.84DD75 pKa = 4.9FKK77 pKa = 11.39CPWAVKK83 pKa = 9.75HH84 pKa = 6.03LSVSQRR90 pKa = 11.84DD91 pKa = 3.77VHH93 pKa = 6.07VAEE96 pKa = 4.43YY97 pKa = 10.86LGLTPVSTVIDD108 pKa = 3.48IDD110 pKa = 3.27QSRR113 pKa = 11.84FDD115 pKa = 3.46RR116 pKa = 11.84HH117 pKa = 6.09ISRR120 pKa = 11.84FWLYY124 pKa = 11.38VEE126 pKa = 4.73TEE128 pKa = 4.08LLSYY132 pKa = 9.39MCHH135 pKa = 6.54PDD137 pKa = 3.42DD138 pKa = 4.42RR139 pKa = 11.84QEE141 pKa = 5.35LRR143 pKa = 11.84ALLNKK148 pKa = 10.27QIVTRR153 pKa = 11.84GFHH156 pKa = 5.04TQGIRR161 pKa = 11.84YY162 pKa = 6.94STVGGRR168 pKa = 11.84CSGDD172 pKa = 3.13ANTSIGNTILNCLITQLAFCSLGYY196 pKa = 9.5TDD198 pKa = 5.91FKK200 pKa = 11.17SVHH203 pKa = 6.91DD204 pKa = 4.81GDD206 pKa = 5.45DD207 pKa = 3.71GLITIYY213 pKa = 10.69GPIMRR218 pKa = 11.84VEE220 pKa = 4.15NNHH223 pKa = 6.13ALEE226 pKa = 4.32SLVEE230 pKa = 4.31DD231 pKa = 3.42LTEE234 pKa = 3.94AMRR237 pKa = 11.84VLGFDD242 pKa = 3.58AKK244 pKa = 10.6IKK246 pKa = 9.31IHH248 pKa = 6.78HH249 pKa = 7.54DD250 pKa = 2.4IHH252 pKa = 6.16TATFDD257 pKa = 3.4GRR259 pKa = 11.84MLYY262 pKa = 10.2FDD264 pKa = 3.98RR265 pKa = 11.84GEE267 pKa = 4.33VKK269 pKa = 10.87SMADD273 pKa = 3.43PLRR276 pKa = 11.84TIVKK280 pKa = 8.34MHH282 pKa = 5.82MTAHH286 pKa = 6.52PTQEE290 pKa = 3.97KK291 pKa = 10.41DD292 pKa = 2.94WTVKK296 pKa = 10.24LRR298 pKa = 11.84NSLLLAKK305 pKa = 9.76ALSYY309 pKa = 10.57AASDD313 pKa = 4.0LDD315 pKa = 3.97TPIIGAWCQFVIRR328 pKa = 11.84NLTHH332 pKa = 6.31NVSHH336 pKa = 7.35PLSKK340 pKa = 10.69SNKK343 pKa = 8.75KK344 pKa = 9.01MLTPRR349 pKa = 11.84YY350 pKa = 8.22PKK352 pKa = 9.93EE353 pKa = 3.6IKK355 pKa = 10.05RR356 pKa = 11.84RR357 pKa = 11.84MDD359 pKa = 4.06LAPIPKK365 pKa = 9.82GYY367 pKa = 10.48LWALRR372 pKa = 11.84KK373 pKa = 10.03RR374 pKa = 11.84NTTIEE379 pKa = 3.51ARR381 pKa = 11.84AAFFVNSNIGTIEE394 pKa = 3.94QEE396 pKa = 4.22MMEE399 pKa = 4.08VWLDD403 pKa = 3.2GSNNIDD409 pKa = 4.39HH410 pKa = 7.31IPPKK414 pKa = 9.88LTIAPRR420 pKa = 11.84EE421 pKa = 3.67FDD423 pKa = 5.43LNDD426 pKa = 3.5LYY428 pKa = 11.77YY429 pKa = 10.62FFSSTHH435 pKa = 6.3EE436 pKa = 3.97YY437 pKa = 8.22HH438 pKa = 6.51TLL440 pKa = 3.35
MM1 pKa = 7.74EE2 pKa = 5.01FPTWVARR9 pKa = 11.84YY10 pKa = 7.18TGGRR14 pKa = 11.84RR15 pKa = 11.84KK16 pKa = 9.4QLQRR20 pKa = 11.84EE21 pKa = 4.1FDD23 pKa = 3.37TGHH26 pKa = 6.44WKK28 pKa = 10.46VADD31 pKa = 3.91EE32 pKa = 4.35AASGVKK38 pKa = 10.26SFIKK42 pKa = 10.27IEE44 pKa = 4.23AGKK47 pKa = 8.27PTDD50 pKa = 3.91PRR52 pKa = 11.84NISPRR57 pKa = 11.84PDD59 pKa = 3.08EE60 pKa = 4.02YY61 pKa = 11.14LVRR64 pKa = 11.84LGPIIAGIEE73 pKa = 3.92RR74 pKa = 11.84DD75 pKa = 4.9FKK77 pKa = 11.39CPWAVKK83 pKa = 9.75HH84 pKa = 6.03LSVSQRR90 pKa = 11.84DD91 pKa = 3.77VHH93 pKa = 6.07VAEE96 pKa = 4.43YY97 pKa = 10.86LGLTPVSTVIDD108 pKa = 3.48IDD110 pKa = 3.27QSRR113 pKa = 11.84FDD115 pKa = 3.46RR116 pKa = 11.84HH117 pKa = 6.09ISRR120 pKa = 11.84FWLYY124 pKa = 11.38VEE126 pKa = 4.73TEE128 pKa = 4.08LLSYY132 pKa = 9.39MCHH135 pKa = 6.54PDD137 pKa = 3.42DD138 pKa = 4.42RR139 pKa = 11.84QEE141 pKa = 5.35LRR143 pKa = 11.84ALLNKK148 pKa = 10.27QIVTRR153 pKa = 11.84GFHH156 pKa = 5.04TQGIRR161 pKa = 11.84YY162 pKa = 6.94STVGGRR168 pKa = 11.84CSGDD172 pKa = 3.13ANTSIGNTILNCLITQLAFCSLGYY196 pKa = 9.5TDD198 pKa = 5.91FKK200 pKa = 11.17SVHH203 pKa = 6.91DD204 pKa = 4.81GDD206 pKa = 5.45DD207 pKa = 3.71GLITIYY213 pKa = 10.69GPIMRR218 pKa = 11.84VEE220 pKa = 4.15NNHH223 pKa = 6.13ALEE226 pKa = 4.32SLVEE230 pKa = 4.31DD231 pKa = 3.42LTEE234 pKa = 3.94AMRR237 pKa = 11.84VLGFDD242 pKa = 3.58AKK244 pKa = 10.6IKK246 pKa = 9.31IHH248 pKa = 6.78HH249 pKa = 7.54DD250 pKa = 2.4IHH252 pKa = 6.16TATFDD257 pKa = 3.4GRR259 pKa = 11.84MLYY262 pKa = 10.2FDD264 pKa = 3.98RR265 pKa = 11.84GEE267 pKa = 4.33VKK269 pKa = 10.87SMADD273 pKa = 3.43PLRR276 pKa = 11.84TIVKK280 pKa = 8.34MHH282 pKa = 5.82MTAHH286 pKa = 6.52PTQEE290 pKa = 3.97KK291 pKa = 10.41DD292 pKa = 2.94WTVKK296 pKa = 10.24LRR298 pKa = 11.84NSLLLAKK305 pKa = 9.76ALSYY309 pKa = 10.57AASDD313 pKa = 4.0LDD315 pKa = 3.97TPIIGAWCQFVIRR328 pKa = 11.84NLTHH332 pKa = 6.31NVSHH336 pKa = 7.35PLSKK340 pKa = 10.69SNKK343 pKa = 8.75KK344 pKa = 9.01MLTPRR349 pKa = 11.84YY350 pKa = 8.22PKK352 pKa = 9.93EE353 pKa = 3.6IKK355 pKa = 10.05RR356 pKa = 11.84RR357 pKa = 11.84MDD359 pKa = 4.06LAPIPKK365 pKa = 9.82GYY367 pKa = 10.48LWALRR372 pKa = 11.84KK373 pKa = 10.03RR374 pKa = 11.84NTTIEE379 pKa = 3.51ARR381 pKa = 11.84AAFFVNSNIGTIEE394 pKa = 3.94QEE396 pKa = 4.22MMEE399 pKa = 4.08VWLDD403 pKa = 3.2GSNNIDD409 pKa = 4.39HH410 pKa = 7.31IPPKK414 pKa = 9.88LTIAPRR420 pKa = 11.84EE421 pKa = 3.67FDD423 pKa = 5.43LNDD426 pKa = 3.5LYY428 pKa = 11.77YY429 pKa = 10.62FFSSTHH435 pKa = 6.3EE436 pKa = 3.97YY437 pKa = 8.22HH438 pKa = 6.51TLL440 pKa = 3.35
Molecular weight: 50.34 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2038 |
364 |
1234 |
679.3 |
75.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.458 ± 0.724 | 1.766 ± 0.152 |
5.545 ± 0.424 | 5.643 ± 0.365 |
3.729 ± 0.138 | 6.771 ± 0.488 |
2.502 ± 0.713 | 5.005 ± 0.788 |
4.269 ± 0.405 | 8.096 ± 0.397 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.846 ± 0.132 | 3.729 ± 0.796 |
5.348 ± 0.55 | 2.846 ± 0.11 |
6.084 ± 0.788 | 8.342 ± 0.768 |
7.066 ± 1.013 | 8.096 ± 0.903 |
1.276 ± 0.186 | 3.582 ± 0.081 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
