
Clostridium taeniosporum
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Eubacteriales; Clostridiaceae; Clostridium
Average proteome isoelectric point is 6.59
Get precalculated fractions of proteins
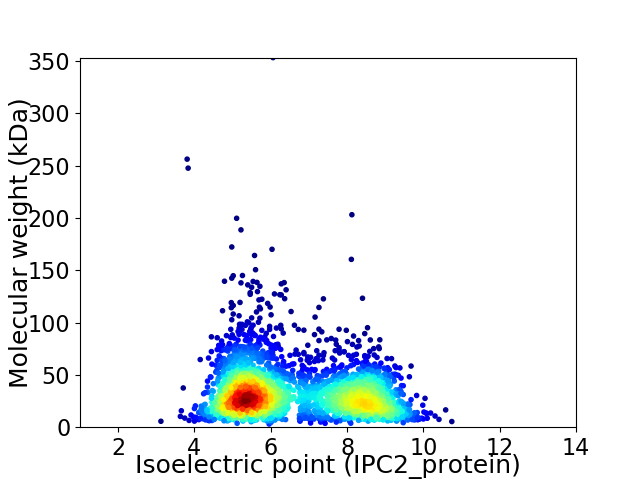
Virtual 2D-PAGE plot for 2948 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1D7XJ94|A0A1D7XJ94_9CLOT NAD-dependent dehydratase OS=Clostridium taeniosporum OX=394958 GN=BGI42_06490 PE=4 SV=1
MM1 pKa = 7.68PSILRR6 pKa = 11.84YY7 pKa = 8.58STTARR12 pKa = 11.84GSMVITGNTLGLLGKK27 pKa = 9.53FNPPDD32 pKa = 3.61ATDD35 pKa = 3.33TDD37 pKa = 4.14VNANGIGAFISLNNGISAPGGYY59 pKa = 9.44PLGTTFDD66 pKa = 3.37WRR68 pKa = 11.84EE69 pKa = 3.87NGSDD73 pKa = 4.11AIIDD77 pKa = 3.63IPVSAKK83 pKa = 9.84ILYY86 pKa = 10.41AEE88 pKa = 4.78LSWSGSYY95 pKa = 11.01LNADD99 pKa = 3.21QDD101 pKa = 3.8VSAFLDD107 pKa = 3.41NAITFNTPLGSFQVNPDD124 pKa = 3.12AATAGSVVFPTNGVGQYY141 pKa = 9.62FRR143 pKa = 11.84SANVTTLVNSGTGTYY158 pKa = 9.77SVQSIPGVLGGTPSNPAVEE177 pKa = 4.18NNYY180 pKa = 10.36LGWSLEE186 pKa = 3.74IAYY189 pKa = 10.05EE190 pKa = 4.31DD191 pKa = 3.18VTLPYY196 pKa = 10.94RR197 pKa = 11.84NMNVWTIFDD206 pKa = 4.38PVDD209 pKa = 3.56LAISPIVDD217 pKa = 3.2IPVSGFSTPISGSIKK232 pKa = 10.36ARR234 pKa = 11.84LLISAGEE241 pKa = 4.22GDD243 pKa = 3.96PQVTGDD249 pKa = 3.65QVLFGPDD256 pKa = 2.77IVGLVALQGEE266 pKa = 4.86PPSPPNNPVDD276 pKa = 3.42NFFVSRR282 pKa = 11.84INDD285 pKa = 3.64GNSEE289 pKa = 4.43SLTVGQVDD297 pKa = 3.78TLGSFGTLNQPVPTTLAGRR316 pKa = 11.84LAPGRR321 pKa = 11.84YY322 pKa = 9.76GYY324 pKa = 10.68DD325 pKa = 2.81ITNVDD330 pKa = 3.56ASTTLVNSQTNALIRR345 pKa = 11.84LTTSGDD351 pKa = 3.6GYY353 pKa = 11.43VPNLLGLQIDD363 pKa = 4.25SNSPIISINKK373 pKa = 8.57SAPATATRR381 pKa = 11.84GSDD384 pKa = 2.66ITYY387 pKa = 9.23TLTVTNTGVIQADD400 pKa = 4.11TVTVLDD406 pKa = 4.42PAPTGTTVNPGSVTVQNATGPVVNNSTAATLNVDD440 pKa = 4.58IGPLLPNQVVTIQYY454 pKa = 7.68TVSTTGTTPTPVLNSTTANYY474 pKa = 9.36TFVPIPGGNVITDD487 pKa = 3.76TSNSNEE493 pKa = 4.4TITAIANKK501 pKa = 9.0FVCEE505 pKa = 3.89PFGYY509 pKa = 9.64RR510 pKa = 11.84VAAEE514 pKa = 4.14STGTNSTLLKK524 pKa = 10.48LNLVTGEE531 pKa = 4.12VDD533 pKa = 3.94VLNPDD538 pKa = 2.53IGMNINAIGYY548 pKa = 8.52NVLDD552 pKa = 3.58NLIYY556 pKa = 10.49GIEE559 pKa = 4.05NTTTNLVILDD569 pKa = 3.93STGTTSVLGPVPNLPLLPSPFYY591 pKa = 10.26NTGTIDD597 pKa = 3.92NNGHH601 pKa = 7.2LYY603 pKa = 10.08IKK605 pKa = 10.67SSTDD609 pKa = 3.01PNYY612 pKa = 10.93YY613 pKa = 10.29VVDD616 pKa = 4.23VNSSSPTFGQLVDD629 pKa = 3.99PLNGFVLDD637 pKa = 3.96TAPYY641 pKa = 7.3GTPINPNMLVADD653 pKa = 3.96WTVSSVDD660 pKa = 3.13GNLYY664 pKa = 10.48GVEE667 pKa = 4.08NTSGNVVSLNPLTGVVTVLTTSGIPIGAYY696 pKa = 9.17GAVYY700 pKa = 10.22SINDD704 pKa = 3.27QYY706 pKa = 11.59IYY708 pKa = 10.36ATNNTTGVIYY718 pKa = 10.41RR719 pKa = 11.84IEE721 pKa = 4.15INGDD725 pKa = 3.03NATGIVFSSDD735 pKa = 3.12VPSNNNDD742 pKa = 2.87GAACLNAEE750 pKa = 4.36LLIDD754 pKa = 5.3FGDD757 pKa = 4.52APDD760 pKa = 4.11PTPGNGPNDD769 pKa = 3.85YY770 pKa = 9.78STLLANNGPRR780 pKa = 11.84HH781 pKa = 5.55QIINQLYY788 pKa = 10.34LGTQVTAEE796 pKa = 3.76NDD798 pKa = 2.99AYY800 pKa = 11.07ANITSDD806 pKa = 3.13ATGDD810 pKa = 4.45DD811 pKa = 4.51IIQGIQDD818 pKa = 4.21DD819 pKa = 4.71GLVVPLTPMLSTDD832 pKa = 2.86TTYY835 pKa = 11.88ALNVSVTNNTGLPANLYY852 pKa = 9.92GWIDD856 pKa = 3.78FNEE859 pKa = 4.33DD860 pKa = 3.77GIFQGNEE867 pKa = 3.41AAPVQIVPSALGLQQFILNFIVPVGVVLQPDD898 pKa = 3.52HH899 pKa = 6.1TFVRR903 pKa = 11.84LRR905 pKa = 11.84LTTDD909 pKa = 3.36DD910 pKa = 5.96LINQNSLPIDD920 pKa = 3.77EE921 pKa = 5.05DD922 pKa = 3.79TRR924 pKa = 11.84SIGPASDD931 pKa = 4.48GEE933 pKa = 4.37VEE935 pKa = 5.3DD936 pKa = 4.9YY937 pKa = 11.06YY938 pKa = 11.37LTITPVADD946 pKa = 2.95ISVVKK951 pKa = 9.79TGAPNPVVVGNTLTYY966 pKa = 10.48TMVVSNLGPSDD977 pKa = 3.77AQSVSLTDD985 pKa = 4.01AVPACILNPVYY996 pKa = 10.52SVNGGTSNPWPVGGINLGTIASGATPITVTISGTVDD1032 pKa = 3.61PACTEE1037 pKa = 3.91SSLVNTATVSSPTPDD1052 pKa = 4.27PDD1054 pKa = 3.74PSNNTSTVTTQVNQSADD1071 pKa = 3.0IKK1073 pKa = 9.93VVKK1076 pKa = 8.71TASPSPVVAGQTLTYY1091 pKa = 10.07TMVVSNLGPSDD1102 pKa = 3.77AQSVSLTDD1110 pKa = 4.01AVPACILNPVYY1121 pKa = 10.54SVNGGASNPWPVGGINLGTIAVGATPITVTISGTVDD1157 pKa = 3.52PACAEE1162 pKa = 4.02SSLVNTAIVSSPTPDD1177 pKa = 4.24PDD1179 pKa = 3.74PSNNTSTVTTQVNQSADD1196 pKa = 2.96ISVVKK1201 pKa = 9.65TGSSNPVVAGNILTYY1216 pKa = 10.79TMVVSNLGPSDD1227 pKa = 3.77AQSVSLTDD1235 pKa = 4.01AVPACILNPVYY1246 pKa = 10.52SVNGGTSNPWPVGGINLGTIASGATPITVTISGTVDD1282 pKa = 3.61PACTEE1287 pKa = 3.91SSLVNTATVSSPTPDD1302 pKa = 4.2LDD1304 pKa = 4.15PNNNTSTVTTTVNTLADD1321 pKa = 3.52VSVVKK1326 pKa = 9.09TANPINVNRR1335 pKa = 11.84GDD1337 pKa = 3.66VVTYY1341 pKa = 8.51TIVVSNAGPSDD1352 pKa = 3.7AQSVSLADD1360 pKa = 3.64VVPSEE1365 pKa = 4.15LLNPEE1370 pKa = 4.06FSINNGVSWNIWSNPYY1386 pKa = 10.65SIGTLTAGTSKK1397 pKa = 9.98TIFIRR1402 pKa = 11.84GTISNTAIGTVNNTATVSSSTPDD1425 pKa = 3.61PNPDD1429 pKa = 3.44NNTSSASIEE1438 pKa = 4.04VNYY1441 pKa = 10.79ADD1443 pKa = 5.44LVAPGNFVKK1452 pKa = 10.71AVDD1455 pKa = 3.87KK1456 pKa = 10.97QYY1458 pKa = 11.84ADD1460 pKa = 3.23IGDD1463 pKa = 4.53EE1464 pKa = 3.59ITYY1467 pKa = 8.5TINVTNTGNTIANNVVITDD1486 pKa = 5.09PIPNGTSYY1494 pKa = 11.63VSGSLIASAPVTGTPAGGINVTNGIAPGATLTLSYY1529 pKa = 9.89KK1530 pKa = 10.5VKK1532 pKa = 10.12VIQMPVPNPIPNTASISYY1550 pKa = 8.94TYY1552 pKa = 10.36TVDD1555 pKa = 3.47PNNPNGASGSGDD1567 pKa = 3.34TNTVFTQVNHH1577 pKa = 6.77GEE1579 pKa = 4.18ILPEE1583 pKa = 4.06NAVKK1587 pKa = 9.41TVDD1590 pKa = 3.95TNPTTPGDD1598 pKa = 3.77IITYY1602 pKa = 7.38TVSIKK1607 pKa = 9.66NTGNVPVDD1615 pKa = 3.5ALVTDD1620 pKa = 4.22VVPTGTQFVTGTVIINGVSKK1640 pKa = 9.96PAKK1643 pKa = 9.84NPNFGIKK1650 pKa = 9.86IIGIPAGEE1658 pKa = 3.96IANLSFKK1665 pKa = 11.21VKK1667 pKa = 10.63VLDD1670 pKa = 3.97NAPSTLINNAKK1681 pKa = 9.86INYY1684 pKa = 9.22SYY1686 pKa = 11.47VVDD1689 pKa = 3.9PALPPVNKK1697 pKa = 10.15DD1698 pKa = 3.04VTTNTVNVTVLKK1710 pKa = 10.87PEE1712 pKa = 3.96LTLVKK1717 pKa = 10.56SADD1720 pKa = 3.57RR1721 pKa = 11.84TGAVVGDD1728 pKa = 4.05IIRR1731 pKa = 11.84YY1732 pKa = 9.84SITATNTGEE1741 pKa = 4.17IPLQNVIVKK1750 pKa = 10.36DD1751 pKa = 3.6VLAPEE1756 pKa = 4.56LSYY1759 pKa = 10.95IGNLTINGVPSGQSILTGVNIEE1781 pKa = 4.48DD1782 pKa = 3.9LSIGEE1787 pKa = 4.35VKK1789 pKa = 10.08TISFDD1794 pKa = 3.55AKK1796 pKa = 10.82VNAIPADD1803 pKa = 3.5KK1804 pKa = 10.23TINNTSTATFNYY1816 pKa = 9.27IVSGKK1821 pKa = 9.47NFSGNATSNEE1831 pKa = 3.79EE1832 pKa = 4.19TIRR1835 pKa = 11.84VYY1837 pKa = 10.97NPEE1840 pKa = 3.61LTMTKK1845 pKa = 9.55MSNNPIVKK1853 pKa = 10.3VGDD1856 pKa = 3.53TFQYY1860 pKa = 10.68TITAEE1865 pKa = 4.04NTGDD1869 pKa = 3.68IIINDD1874 pKa = 4.17VIVKK1878 pKa = 10.39DD1879 pKa = 3.86DD1880 pKa = 4.39LPPEE1884 pKa = 4.16FEE1886 pKa = 4.26VVQIIVNGDD1895 pKa = 3.58VVNGDD1900 pKa = 3.11IEE1902 pKa = 4.48AGINIGNLAIGEE1914 pKa = 4.4SVDD1917 pKa = 3.25IVLTIQVLADD1927 pKa = 3.61LTDD1930 pKa = 3.77TFQNIATGTGTSITDD1945 pKa = 3.62PNKK1948 pKa = 10.11PPVIVEE1954 pKa = 4.41GEE1956 pKa = 4.34GEE1958 pKa = 4.18DD1959 pKa = 4.22PGVTVYY1965 pKa = 10.77NPKK1968 pKa = 10.64LEE1970 pKa = 4.31LEE1972 pKa = 4.29KK1973 pKa = 11.09SVDD1976 pKa = 3.29KK1977 pKa = 10.74PYY1979 pKa = 11.05VIVGDD1984 pKa = 3.9TVTYY1988 pKa = 9.41TVVAKK1993 pKa = 9.29NTGDD1997 pKa = 3.25IVLGNVDD2004 pKa = 5.07LDD2006 pKa = 4.36PIVIYY2011 pKa = 10.75DD2012 pKa = 4.02LLGPSLEE2019 pKa = 4.25FVSGSVTIDD2028 pKa = 4.27GISDD2032 pKa = 3.87PLSGIVNGVILGVLNVGEE2050 pKa = 4.53SKK2052 pKa = 9.78TVTFKK2057 pKa = 11.47AKK2059 pKa = 10.3VLSNDD2064 pKa = 2.65ISPIEE2069 pKa = 4.1NTAQATFGYY2078 pKa = 9.31QIPGDD2083 pKa = 3.84EE2084 pKa = 4.74PEE2086 pKa = 4.59TGNATSNTVQVIPEE2100 pKa = 3.97IANLEE2105 pKa = 4.06ILKK2108 pKa = 10.49EE2109 pKa = 3.92ADD2111 pKa = 3.22TNFAVMNDD2119 pKa = 3.64VINYY2123 pKa = 7.03TVTVTNTGTLDD2134 pKa = 3.4ANNVIFTDD2142 pKa = 3.68EE2143 pKa = 4.57LPPEE2147 pKa = 4.24VEE2149 pKa = 5.19LIDD2152 pKa = 4.79GSFKK2156 pKa = 10.95VDD2158 pKa = 4.21GIGINNVDD2166 pKa = 3.65LRR2168 pKa = 11.84QGVNIGTIKK2177 pKa = 9.94TGEE2180 pKa = 3.95EE2181 pKa = 3.89AIIEE2185 pKa = 4.21YY2186 pKa = 9.27SVKK2189 pKa = 10.63VIATNCEE2196 pKa = 3.97LKK2198 pKa = 10.53IKK2200 pKa = 10.57NSAIAKK2206 pKa = 7.4FQYY2209 pKa = 10.26RR2210 pKa = 11.84LPNGTTGTVTTDD2222 pKa = 3.21PSEE2225 pKa = 4.1TSTVIVNLGISNFKK2239 pKa = 10.51QLFIEE2244 pKa = 4.42EE2245 pKa = 4.2NLIIPDD2251 pKa = 4.01EE2252 pKa = 4.64KK2253 pKa = 11.03PDD2255 pKa = 3.37IEE2257 pKa = 5.26EE2258 pKa = 3.99INEE2261 pKa = 3.84VNGDD2265 pKa = 3.55VEE2267 pKa = 4.59IIKK2270 pKa = 10.12HH2271 pKa = 4.96YY2272 pKa = 10.71VVEE2275 pKa = 4.33TPILKK2280 pKa = 10.62SNEE2283 pKa = 3.76GQILTGCKK2291 pKa = 10.22LIIQGVLNEE2300 pKa = 4.26VVEE2303 pKa = 4.37YY2304 pKa = 9.16TALDD2308 pKa = 3.72SEE2310 pKa = 4.62QTIHH2314 pKa = 6.23SAHH2317 pKa = 6.04YY2318 pKa = 9.62SIPFSTFIILPEE2330 pKa = 4.0NYY2332 pKa = 9.7CYY2334 pKa = 11.05GNEE2337 pKa = 4.7DD2338 pKa = 2.94ITTEE2342 pKa = 4.22VEE2344 pKa = 4.36DD2345 pKa = 4.84IYY2347 pKa = 11.4FNSLNSRR2354 pKa = 11.84KK2355 pKa = 9.83LFTNATILIKK2365 pKa = 10.43ATVSYY2370 pKa = 10.71CKK2372 pKa = 10.51
MM1 pKa = 7.68PSILRR6 pKa = 11.84YY7 pKa = 8.58STTARR12 pKa = 11.84GSMVITGNTLGLLGKK27 pKa = 9.53FNPPDD32 pKa = 3.61ATDD35 pKa = 3.33TDD37 pKa = 4.14VNANGIGAFISLNNGISAPGGYY59 pKa = 9.44PLGTTFDD66 pKa = 3.37WRR68 pKa = 11.84EE69 pKa = 3.87NGSDD73 pKa = 4.11AIIDD77 pKa = 3.63IPVSAKK83 pKa = 9.84ILYY86 pKa = 10.41AEE88 pKa = 4.78LSWSGSYY95 pKa = 11.01LNADD99 pKa = 3.21QDD101 pKa = 3.8VSAFLDD107 pKa = 3.41NAITFNTPLGSFQVNPDD124 pKa = 3.12AATAGSVVFPTNGVGQYY141 pKa = 9.62FRR143 pKa = 11.84SANVTTLVNSGTGTYY158 pKa = 9.77SVQSIPGVLGGTPSNPAVEE177 pKa = 4.18NNYY180 pKa = 10.36LGWSLEE186 pKa = 3.74IAYY189 pKa = 10.05EE190 pKa = 4.31DD191 pKa = 3.18VTLPYY196 pKa = 10.94RR197 pKa = 11.84NMNVWTIFDD206 pKa = 4.38PVDD209 pKa = 3.56LAISPIVDD217 pKa = 3.2IPVSGFSTPISGSIKK232 pKa = 10.36ARR234 pKa = 11.84LLISAGEE241 pKa = 4.22GDD243 pKa = 3.96PQVTGDD249 pKa = 3.65QVLFGPDD256 pKa = 2.77IVGLVALQGEE266 pKa = 4.86PPSPPNNPVDD276 pKa = 3.42NFFVSRR282 pKa = 11.84INDD285 pKa = 3.64GNSEE289 pKa = 4.43SLTVGQVDD297 pKa = 3.78TLGSFGTLNQPVPTTLAGRR316 pKa = 11.84LAPGRR321 pKa = 11.84YY322 pKa = 9.76GYY324 pKa = 10.68DD325 pKa = 2.81ITNVDD330 pKa = 3.56ASTTLVNSQTNALIRR345 pKa = 11.84LTTSGDD351 pKa = 3.6GYY353 pKa = 11.43VPNLLGLQIDD363 pKa = 4.25SNSPIISINKK373 pKa = 8.57SAPATATRR381 pKa = 11.84GSDD384 pKa = 2.66ITYY387 pKa = 9.23TLTVTNTGVIQADD400 pKa = 4.11TVTVLDD406 pKa = 4.42PAPTGTTVNPGSVTVQNATGPVVNNSTAATLNVDD440 pKa = 4.58IGPLLPNQVVTIQYY454 pKa = 7.68TVSTTGTTPTPVLNSTTANYY474 pKa = 9.36TFVPIPGGNVITDD487 pKa = 3.76TSNSNEE493 pKa = 4.4TITAIANKK501 pKa = 9.0FVCEE505 pKa = 3.89PFGYY509 pKa = 9.64RR510 pKa = 11.84VAAEE514 pKa = 4.14STGTNSTLLKK524 pKa = 10.48LNLVTGEE531 pKa = 4.12VDD533 pKa = 3.94VLNPDD538 pKa = 2.53IGMNINAIGYY548 pKa = 8.52NVLDD552 pKa = 3.58NLIYY556 pKa = 10.49GIEE559 pKa = 4.05NTTTNLVILDD569 pKa = 3.93STGTTSVLGPVPNLPLLPSPFYY591 pKa = 10.26NTGTIDD597 pKa = 3.92NNGHH601 pKa = 7.2LYY603 pKa = 10.08IKK605 pKa = 10.67SSTDD609 pKa = 3.01PNYY612 pKa = 10.93YY613 pKa = 10.29VVDD616 pKa = 4.23VNSSSPTFGQLVDD629 pKa = 3.99PLNGFVLDD637 pKa = 3.96TAPYY641 pKa = 7.3GTPINPNMLVADD653 pKa = 3.96WTVSSVDD660 pKa = 3.13GNLYY664 pKa = 10.48GVEE667 pKa = 4.08NTSGNVVSLNPLTGVVTVLTTSGIPIGAYY696 pKa = 9.17GAVYY700 pKa = 10.22SINDD704 pKa = 3.27QYY706 pKa = 11.59IYY708 pKa = 10.36ATNNTTGVIYY718 pKa = 10.41RR719 pKa = 11.84IEE721 pKa = 4.15INGDD725 pKa = 3.03NATGIVFSSDD735 pKa = 3.12VPSNNNDD742 pKa = 2.87GAACLNAEE750 pKa = 4.36LLIDD754 pKa = 5.3FGDD757 pKa = 4.52APDD760 pKa = 4.11PTPGNGPNDD769 pKa = 3.85YY770 pKa = 9.78STLLANNGPRR780 pKa = 11.84HH781 pKa = 5.55QIINQLYY788 pKa = 10.34LGTQVTAEE796 pKa = 3.76NDD798 pKa = 2.99AYY800 pKa = 11.07ANITSDD806 pKa = 3.13ATGDD810 pKa = 4.45DD811 pKa = 4.51IIQGIQDD818 pKa = 4.21DD819 pKa = 4.71GLVVPLTPMLSTDD832 pKa = 2.86TTYY835 pKa = 11.88ALNVSVTNNTGLPANLYY852 pKa = 9.92GWIDD856 pKa = 3.78FNEE859 pKa = 4.33DD860 pKa = 3.77GIFQGNEE867 pKa = 3.41AAPVQIVPSALGLQQFILNFIVPVGVVLQPDD898 pKa = 3.52HH899 pKa = 6.1TFVRR903 pKa = 11.84LRR905 pKa = 11.84LTTDD909 pKa = 3.36DD910 pKa = 5.96LINQNSLPIDD920 pKa = 3.77EE921 pKa = 5.05DD922 pKa = 3.79TRR924 pKa = 11.84SIGPASDD931 pKa = 4.48GEE933 pKa = 4.37VEE935 pKa = 5.3DD936 pKa = 4.9YY937 pKa = 11.06YY938 pKa = 11.37LTITPVADD946 pKa = 2.95ISVVKK951 pKa = 9.79TGAPNPVVVGNTLTYY966 pKa = 10.48TMVVSNLGPSDD977 pKa = 3.77AQSVSLTDD985 pKa = 4.01AVPACILNPVYY996 pKa = 10.52SVNGGTSNPWPVGGINLGTIASGATPITVTISGTVDD1032 pKa = 3.61PACTEE1037 pKa = 3.91SSLVNTATVSSPTPDD1052 pKa = 4.27PDD1054 pKa = 3.74PSNNTSTVTTQVNQSADD1071 pKa = 3.0IKK1073 pKa = 9.93VVKK1076 pKa = 8.71TASPSPVVAGQTLTYY1091 pKa = 10.07TMVVSNLGPSDD1102 pKa = 3.77AQSVSLTDD1110 pKa = 4.01AVPACILNPVYY1121 pKa = 10.54SVNGGASNPWPVGGINLGTIAVGATPITVTISGTVDD1157 pKa = 3.52PACAEE1162 pKa = 4.02SSLVNTAIVSSPTPDD1177 pKa = 4.24PDD1179 pKa = 3.74PSNNTSTVTTQVNQSADD1196 pKa = 2.96ISVVKK1201 pKa = 9.65TGSSNPVVAGNILTYY1216 pKa = 10.79TMVVSNLGPSDD1227 pKa = 3.77AQSVSLTDD1235 pKa = 4.01AVPACILNPVYY1246 pKa = 10.52SVNGGTSNPWPVGGINLGTIASGATPITVTISGTVDD1282 pKa = 3.61PACTEE1287 pKa = 3.91SSLVNTATVSSPTPDD1302 pKa = 4.2LDD1304 pKa = 4.15PNNNTSTVTTTVNTLADD1321 pKa = 3.52VSVVKK1326 pKa = 9.09TANPINVNRR1335 pKa = 11.84GDD1337 pKa = 3.66VVTYY1341 pKa = 8.51TIVVSNAGPSDD1352 pKa = 3.7AQSVSLADD1360 pKa = 3.64VVPSEE1365 pKa = 4.15LLNPEE1370 pKa = 4.06FSINNGVSWNIWSNPYY1386 pKa = 10.65SIGTLTAGTSKK1397 pKa = 9.98TIFIRR1402 pKa = 11.84GTISNTAIGTVNNTATVSSSTPDD1425 pKa = 3.61PNPDD1429 pKa = 3.44NNTSSASIEE1438 pKa = 4.04VNYY1441 pKa = 10.79ADD1443 pKa = 5.44LVAPGNFVKK1452 pKa = 10.71AVDD1455 pKa = 3.87KK1456 pKa = 10.97QYY1458 pKa = 11.84ADD1460 pKa = 3.23IGDD1463 pKa = 4.53EE1464 pKa = 3.59ITYY1467 pKa = 8.5TINVTNTGNTIANNVVITDD1486 pKa = 5.09PIPNGTSYY1494 pKa = 11.63VSGSLIASAPVTGTPAGGINVTNGIAPGATLTLSYY1529 pKa = 9.89KK1530 pKa = 10.5VKK1532 pKa = 10.12VIQMPVPNPIPNTASISYY1550 pKa = 8.94TYY1552 pKa = 10.36TVDD1555 pKa = 3.47PNNPNGASGSGDD1567 pKa = 3.34TNTVFTQVNHH1577 pKa = 6.77GEE1579 pKa = 4.18ILPEE1583 pKa = 4.06NAVKK1587 pKa = 9.41TVDD1590 pKa = 3.95TNPTTPGDD1598 pKa = 3.77IITYY1602 pKa = 7.38TVSIKK1607 pKa = 9.66NTGNVPVDD1615 pKa = 3.5ALVTDD1620 pKa = 4.22VVPTGTQFVTGTVIINGVSKK1640 pKa = 9.96PAKK1643 pKa = 9.84NPNFGIKK1650 pKa = 9.86IIGIPAGEE1658 pKa = 3.96IANLSFKK1665 pKa = 11.21VKK1667 pKa = 10.63VLDD1670 pKa = 3.97NAPSTLINNAKK1681 pKa = 9.86INYY1684 pKa = 9.22SYY1686 pKa = 11.47VVDD1689 pKa = 3.9PALPPVNKK1697 pKa = 10.15DD1698 pKa = 3.04VTTNTVNVTVLKK1710 pKa = 10.87PEE1712 pKa = 3.96LTLVKK1717 pKa = 10.56SADD1720 pKa = 3.57RR1721 pKa = 11.84TGAVVGDD1728 pKa = 4.05IIRR1731 pKa = 11.84YY1732 pKa = 9.84SITATNTGEE1741 pKa = 4.17IPLQNVIVKK1750 pKa = 10.36DD1751 pKa = 3.6VLAPEE1756 pKa = 4.56LSYY1759 pKa = 10.95IGNLTINGVPSGQSILTGVNIEE1781 pKa = 4.48DD1782 pKa = 3.9LSIGEE1787 pKa = 4.35VKK1789 pKa = 10.08TISFDD1794 pKa = 3.55AKK1796 pKa = 10.82VNAIPADD1803 pKa = 3.5KK1804 pKa = 10.23TINNTSTATFNYY1816 pKa = 9.27IVSGKK1821 pKa = 9.47NFSGNATSNEE1831 pKa = 3.79EE1832 pKa = 4.19TIRR1835 pKa = 11.84VYY1837 pKa = 10.97NPEE1840 pKa = 3.61LTMTKK1845 pKa = 9.55MSNNPIVKK1853 pKa = 10.3VGDD1856 pKa = 3.53TFQYY1860 pKa = 10.68TITAEE1865 pKa = 4.04NTGDD1869 pKa = 3.68IIINDD1874 pKa = 4.17VIVKK1878 pKa = 10.39DD1879 pKa = 3.86DD1880 pKa = 4.39LPPEE1884 pKa = 4.16FEE1886 pKa = 4.26VVQIIVNGDD1895 pKa = 3.58VVNGDD1900 pKa = 3.11IEE1902 pKa = 4.48AGINIGNLAIGEE1914 pKa = 4.4SVDD1917 pKa = 3.25IVLTIQVLADD1927 pKa = 3.61LTDD1930 pKa = 3.77TFQNIATGTGTSITDD1945 pKa = 3.62PNKK1948 pKa = 10.11PPVIVEE1954 pKa = 4.41GEE1956 pKa = 4.34GEE1958 pKa = 4.18DD1959 pKa = 4.22PGVTVYY1965 pKa = 10.77NPKK1968 pKa = 10.64LEE1970 pKa = 4.31LEE1972 pKa = 4.29KK1973 pKa = 11.09SVDD1976 pKa = 3.29KK1977 pKa = 10.74PYY1979 pKa = 11.05VIVGDD1984 pKa = 3.9TVTYY1988 pKa = 9.41TVVAKK1993 pKa = 9.29NTGDD1997 pKa = 3.25IVLGNVDD2004 pKa = 5.07LDD2006 pKa = 4.36PIVIYY2011 pKa = 10.75DD2012 pKa = 4.02LLGPSLEE2019 pKa = 4.25FVSGSVTIDD2028 pKa = 4.27GISDD2032 pKa = 3.87PLSGIVNGVILGVLNVGEE2050 pKa = 4.53SKK2052 pKa = 9.78TVTFKK2057 pKa = 11.47AKK2059 pKa = 10.3VLSNDD2064 pKa = 2.65ISPIEE2069 pKa = 4.1NTAQATFGYY2078 pKa = 9.31QIPGDD2083 pKa = 3.84EE2084 pKa = 4.74PEE2086 pKa = 4.59TGNATSNTVQVIPEE2100 pKa = 3.97IANLEE2105 pKa = 4.06ILKK2108 pKa = 10.49EE2109 pKa = 3.92ADD2111 pKa = 3.22TNFAVMNDD2119 pKa = 3.64VINYY2123 pKa = 7.03TVTVTNTGTLDD2134 pKa = 3.4ANNVIFTDD2142 pKa = 3.68EE2143 pKa = 4.57LPPEE2147 pKa = 4.24VEE2149 pKa = 5.19LIDD2152 pKa = 4.79GSFKK2156 pKa = 10.95VDD2158 pKa = 4.21GIGINNVDD2166 pKa = 3.65LRR2168 pKa = 11.84QGVNIGTIKK2177 pKa = 9.94TGEE2180 pKa = 3.95EE2181 pKa = 3.89AIIEE2185 pKa = 4.21YY2186 pKa = 9.27SVKK2189 pKa = 10.63VIATNCEE2196 pKa = 3.97LKK2198 pKa = 10.53IKK2200 pKa = 10.57NSAIAKK2206 pKa = 7.4FQYY2209 pKa = 10.26RR2210 pKa = 11.84LPNGTTGTVTTDD2222 pKa = 3.21PSEE2225 pKa = 4.1TSTVIVNLGISNFKK2239 pKa = 10.51QLFIEE2244 pKa = 4.42EE2245 pKa = 4.2NLIIPDD2251 pKa = 4.01EE2252 pKa = 4.64KK2253 pKa = 11.03PDD2255 pKa = 3.37IEE2257 pKa = 5.26EE2258 pKa = 3.99INEE2261 pKa = 3.84VNGDD2265 pKa = 3.55VEE2267 pKa = 4.59IIKK2270 pKa = 10.12HH2271 pKa = 4.96YY2272 pKa = 10.71VVEE2275 pKa = 4.33TPILKK2280 pKa = 10.62SNEE2283 pKa = 3.76GQILTGCKK2291 pKa = 10.22LIIQGVLNEE2300 pKa = 4.26VVEE2303 pKa = 4.37YY2304 pKa = 9.16TALDD2308 pKa = 3.72SEE2310 pKa = 4.62QTIHH2314 pKa = 6.23SAHH2317 pKa = 6.04YY2318 pKa = 9.62SIPFSTFIILPEE2330 pKa = 4.0NYY2332 pKa = 9.7CYY2334 pKa = 11.05GNEE2337 pKa = 4.7DD2338 pKa = 2.94ITTEE2342 pKa = 4.22VEE2344 pKa = 4.36DD2345 pKa = 4.84IYY2347 pKa = 11.4FNSLNSRR2354 pKa = 11.84KK2355 pKa = 9.83LFTNATILIKK2365 pKa = 10.43ATVSYY2370 pKa = 10.71CKK2372 pKa = 10.51
Molecular weight: 247.6 kDa
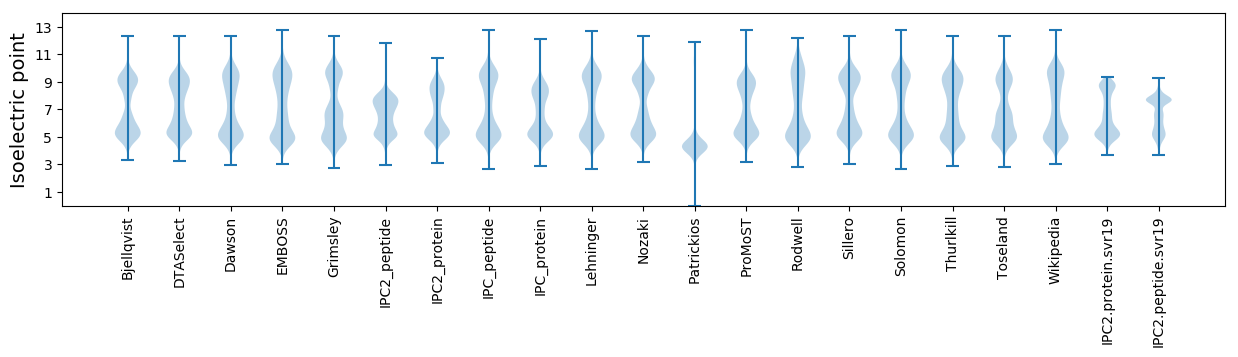
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1D7XNH1|A0A1D7XNH1_9CLOT HEPN domain-containing protein OS=Clostridium taeniosporum OX=394958 GN=BGI42_13800 PE=4 SV=1
MM1 pKa = 7.17GRR3 pKa = 11.84EE4 pKa = 4.13DD5 pKa = 3.34NRR7 pKa = 11.84RR8 pKa = 11.84SGGRR12 pKa = 11.84GRR14 pKa = 11.84RR15 pKa = 11.84SKK17 pKa = 10.92RR18 pKa = 11.84KK19 pKa = 8.67VCAFCVDD26 pKa = 3.31KK27 pKa = 11.49AEE29 pKa = 4.67SIDD32 pKa = 3.62YY33 pKa = 10.94KK34 pKa = 11.04DD35 pKa = 3.2INKK38 pKa = 9.06LRR40 pKa = 11.84KK41 pKa = 9.21YY42 pKa = 7.83VTEE45 pKa = 4.22RR46 pKa = 11.84GKK48 pKa = 10.15ILPRR52 pKa = 11.84RR53 pKa = 11.84ISGTCAKK60 pKa = 9.97HH61 pKa = 5.6QRR63 pKa = 11.84QLTDD67 pKa = 4.19AIKK70 pKa = 10.18RR71 pKa = 11.84ARR73 pKa = 11.84NIALLPFTTEE83 pKa = 3.59
MM1 pKa = 7.17GRR3 pKa = 11.84EE4 pKa = 4.13DD5 pKa = 3.34NRR7 pKa = 11.84RR8 pKa = 11.84SGGRR12 pKa = 11.84GRR14 pKa = 11.84RR15 pKa = 11.84SKK17 pKa = 10.92RR18 pKa = 11.84KK19 pKa = 8.67VCAFCVDD26 pKa = 3.31KK27 pKa = 11.49AEE29 pKa = 4.67SIDD32 pKa = 3.62YY33 pKa = 10.94KK34 pKa = 11.04DD35 pKa = 3.2INKK38 pKa = 9.06LRR40 pKa = 11.84KK41 pKa = 9.21YY42 pKa = 7.83VTEE45 pKa = 4.22RR46 pKa = 11.84GKK48 pKa = 10.15ILPRR52 pKa = 11.84RR53 pKa = 11.84ISGTCAKK60 pKa = 9.97HH61 pKa = 5.6QRR63 pKa = 11.84QLTDD67 pKa = 4.19AIKK70 pKa = 10.18RR71 pKa = 11.84ARR73 pKa = 11.84NIALLPFTTEE83 pKa = 3.59
Molecular weight: 9.61 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
916512 |
26 |
3051 |
310.9 |
35.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.084 ± 0.051 | 1.233 ± 0.018 |
5.699 ± 0.033 | 7.495 ± 0.051 |
4.366 ± 0.034 | 6.164 ± 0.045 |
1.25 ± 0.016 | 10.383 ± 0.064 |
9.691 ± 0.051 | 8.998 ± 0.047 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.459 ± 0.019 | 7.133 ± 0.049 |
2.605 ± 0.03 | 2.187 ± 0.019 |
3.103 ± 0.031 | 6.325 ± 0.038 |
4.827 ± 0.049 | 6.14 ± 0.043 |
0.672 ± 0.014 | 4.187 ± 0.036 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
