
Setaria viridis (Green bristlegrass) (Setaria italica subsp. viridis)
Taxonomy: cellular organisms; Eukaryota; Viridiplantae; Streptophyta; Streptophytina; Embryophyta; Tracheophyta; Euphyllophyta; Spermatophyta; Magnoliopsida; Mesangiospermae; Liliopsida; Petrosaviidae; commelinids; Poales; Poaceae; PACMAD clade; Panicoideae; Panicodae; Paniceae; Cenchrinae; Setaria
Average proteome isoelectric point is 6.98
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 46514 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
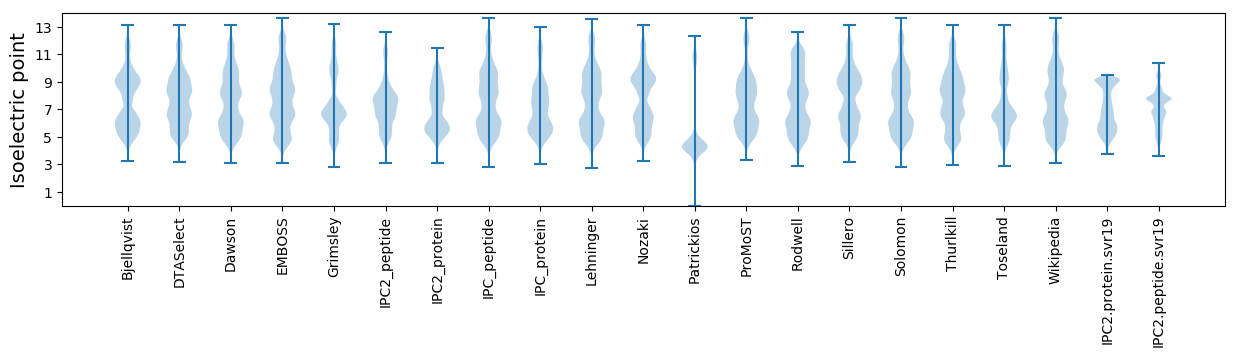
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4U6U9S9|A0A4U6U9S9_SETVI Uncharacterized protein OS=Setaria viridis OX=4556 GN=SEVIR_6G230750v2 PE=4 SV=1
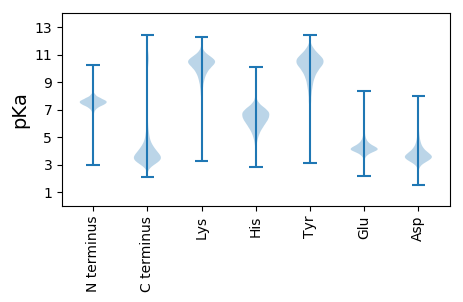
MM1 pKa = 7.49GSDD4 pKa = 3.52FFVFLTGCFTFGCLGLEE21 pKa = 4.27GGARR25 pKa = 11.84GFLRR29 pKa = 11.84GVAGSLLVGGLSPSDD44 pKa = 3.79GDD46 pKa = 3.59WEE48 pKa = 4.56GVLWSSSLAPLGFYY62 pKa = 10.89GDD64 pKa = 4.59GYY66 pKa = 10.77DD67 pKa = 5.12DD68 pKa = 4.87QGGCDD73 pKa = 4.22GPVGDD78 pKa = 5.19DD79 pKa = 3.91GPRR82 pKa = 11.84GGCAGDD88 pKa = 4.67DD89 pKa = 3.54GLKK92 pKa = 10.38FF93 pKa = 3.58
MM1 pKa = 7.49GSDD4 pKa = 3.52FFVFLTGCFTFGCLGLEE21 pKa = 4.27GGARR25 pKa = 11.84GFLRR29 pKa = 11.84GVAGSLLVGGLSPSDD44 pKa = 3.79GDD46 pKa = 3.59WEE48 pKa = 4.56GVLWSSSLAPLGFYY62 pKa = 10.89GDD64 pKa = 4.59GYY66 pKa = 10.77DD67 pKa = 5.12DD68 pKa = 4.87QGGCDD73 pKa = 4.22GPVGDD78 pKa = 5.19DD79 pKa = 3.91GPRR82 pKa = 11.84GGCAGDD88 pKa = 4.67DD89 pKa = 3.54GLKK92 pKa = 10.38FF93 pKa = 3.58
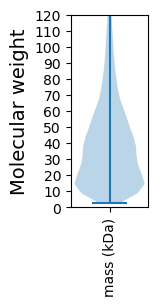
Molecular weight: 9.34 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4U6TPC2|A0A4U6TPC2_SETVI Uncharacterized protein OS=Setaria viridis OX=4556 GN=SEVIR_7G003400v2 PE=4 SV=1
MM1 pKa = 7.85ASRR4 pKa = 11.84SITYY8 pKa = 9.4PGLLLLVVALVIVPSSQAKK27 pKa = 9.64RR28 pKa = 11.84SPPRR32 pKa = 11.84SAPTPAPRR40 pKa = 11.84TAPSPAPRR48 pKa = 11.84SAPTPAPRR56 pKa = 11.84SAPTPAPRR64 pKa = 11.84APQAPSPTPAPTPAPRR80 pKa = 11.84TAPTPAPRR88 pKa = 11.84SAPTPAPRR96 pKa = 11.84APQAPSPTPAPKK108 pKa = 9.98PPPRR112 pKa = 11.84TAPTTAPTPAPRR124 pKa = 11.84SAPTPAPRR132 pKa = 11.84APQAPSPTPAPTPAPTPAPRR152 pKa = 11.84TAPTPAPRR160 pKa = 11.84SAPTPAPRR168 pKa = 11.84APQAPSPTPAPMPAPRR184 pKa = 11.84TAPTLAPRR192 pKa = 11.84SAPTPAPRR200 pKa = 11.84APQAPSPTPAPTPAPRR216 pKa = 11.84TAPISAPTPAPRR228 pKa = 11.84SAPTPTPRR236 pKa = 11.84APQAPSPTPAPTPAPRR252 pKa = 11.84TAPTQAPTPAPRR264 pKa = 11.84SAPTPAPRR272 pKa = 11.84APQVPSPTPAPTPAPRR288 pKa = 11.84TAPTPAPTPAPRR300 pKa = 11.84STPTPAPRR308 pKa = 11.84APQAPSPTPAPTPAPRR324 pKa = 11.84TAPTPAPTPTPRR336 pKa = 11.84SAPTPAPRR344 pKa = 11.84APQVPSPTPTPTPAPRR360 pKa = 11.84TAPTPAPRR368 pKa = 11.84SAPTPAPRR376 pKa = 11.84APQAPSPTPAPTPAPRR392 pKa = 11.84TAPTPAPTPAPRR404 pKa = 11.84SAPTPAPRR412 pKa = 11.84APQAPSPTPTPTPAPRR428 pKa = 11.84TAPTPAPTPAPTPAPRR444 pKa = 11.84SAPTPAPRR452 pKa = 11.84APQAPSPTPAPTPAPRR468 pKa = 11.84TAPTPAPRR476 pKa = 11.84SAPTPAPRR484 pKa = 11.84APQAPSPTPTPTPAPRR500 pKa = 11.84TAPTPAPTPAPTPAPRR516 pKa = 11.84SAPTPAPRR524 pKa = 11.84APQAPSPTPAPTPAPRR540 pKa = 11.84TAPTPAPRR548 pKa = 11.84SAPTPAPRR556 pKa = 11.84APQAPSPTPAPTPAPRR572 pKa = 11.84TAPTPAPRR580 pKa = 11.84SAPTPAPRR588 pKa = 11.84APQAPSPTPAPTPAPRR604 pKa = 11.84TAPTPAPRR612 pKa = 11.84SAPTPAPRR620 pKa = 11.84APQAPSPTPAPTPAPRR636 pKa = 11.84MAPTPASTPAPRR648 pKa = 11.84SAPTPAPRR656 pKa = 11.84ASQAPSPTPAPTPAPRR672 pKa = 11.84MAPTPAPRR680 pKa = 11.84SVPTPAPRR688 pKa = 11.84APLAPSPTPAPTLAPRR704 pKa = 11.84TAPTPAPRR712 pKa = 11.84SAPTLAPRR720 pKa = 11.84APQAPSPRR728 pKa = 11.84PAPTPAPRR736 pKa = 11.84TAPTPAPTPTPRR748 pKa = 11.84SAPSPAPRR756 pKa = 11.84APQVPSPTRR765 pKa = 11.84APTPAPRR772 pKa = 11.84TAPTMAPTPAPRR784 pKa = 11.84SAPTPAPRR792 pKa = 11.84APQAPSPTPAPTPAPRR808 pKa = 11.84TAPTPAPTPAPRR820 pKa = 11.84SAPTPAPRR828 pKa = 11.84APQAPSPTHH837 pKa = 6.79APMLAPRR844 pKa = 11.84TAPTPAPTPTPRR856 pKa = 11.84LAPTPAPRR864 pKa = 11.84APQAPSPTTTPTPAPRR880 pKa = 11.84TAPTPTPRR888 pKa = 11.84SAPTPAPRR896 pKa = 11.84APQAPSPTPAPTQAPRR912 pKa = 11.84TAPTPAPTPAPRR924 pKa = 11.84SAPTPAPRR932 pKa = 11.84APQAPSPTPAPTPAPRR948 pKa = 11.84TAPTPAPTPAPRR960 pKa = 11.84SAPTPAPRR968 pKa = 11.84PPQAPSPTPAPTPASRR984 pKa = 11.84TAPTPSPRR992 pKa = 11.84SAPTPAPRR1000 pKa = 11.84APLAPSPTPAPTPAPRR1016 pKa = 11.84TAPTPAPRR1024 pKa = 11.84SAPTPAPRR1032 pKa = 11.84APQAPSPTPAPTPAPRR1048 pKa = 11.84TAPTPAPRR1056 pKa = 11.84SAPTPTPRR1064 pKa = 11.84APQAPSPTPAPTPAPRR1080 pKa = 11.84MAPTPAPTPAPRR1092 pKa = 11.84SAPTPAPRR1100 pKa = 11.84ASQAPSPTPAPTPAPRR1116 pKa = 11.84MAPTPAPRR1124 pKa = 11.84SVPTPAPRR1132 pKa = 11.84APLAPSPTPAPTLAPRR1148 pKa = 11.84TAPTPAPRR1156 pKa = 11.84SAPTLAPRR1164 pKa = 11.84APQAPSPTPAPTPAPRR1180 pKa = 11.84TAPTPAPTPTPRR1192 pKa = 11.84SAPSPAPRR1200 pKa = 11.84APQVPSPTPAPTPAPRR1216 pKa = 11.84TAPTTAPTPAPRR1228 pKa = 11.84SAPTPAPRR1236 pKa = 11.84APQAPSPTPAPTPAPRR1252 pKa = 11.84TAPTPAPTPTPRR1264 pKa = 11.84SAPTPAPRR1272 pKa = 11.84APQAPSPTPAPMLAPRR1288 pKa = 11.84TAPTPAPTPTPRR1300 pKa = 11.84LAPTPAPRR1308 pKa = 11.84APQAPSPTPTPTPAPRR1324 pKa = 11.84MAPTPAPRR1332 pKa = 11.84SAPTPAPRR1340 pKa = 11.84APQAPSPTPAPTQAPRR1356 pKa = 11.84TAPTPAPTPAPRR1368 pKa = 11.84SAPTPAPRR1376 pKa = 11.84APQAPSPTPAPTPAPRR1392 pKa = 11.84TAPTPAPTPAPRR1404 pKa = 11.84SAPTPTPRR1412 pKa = 11.84PPQAPSPTPAPTPASRR1428 pKa = 11.84TAPTPAPRR1436 pKa = 11.84SAPTPAPRR1444 pKa = 11.84APQAPSPTPAPRR1456 pKa = 11.84TAPTPAPRR1464 pKa = 11.84SAPTPAPRR1472 pKa = 11.84APQAPSPTPAPTPAPRR1488 pKa = 11.84TAPTPAPTPAPRR1500 pKa = 11.84FAPTPAPRR1508 pKa = 11.84APQAPSPTPAPTPAPRR1524 pKa = 11.84TAPTPAPTPAPRR1536 pKa = 11.84SAPTLAPRR1544 pKa = 11.84APQAPSPTPTPTPAPRR1560 pKa = 11.84TAPTPAPTPAPRR1572 pKa = 11.84SAPTPAPRR1580 pKa = 11.84APQAPSPTPAPTPAPRR1596 pKa = 11.84TAPTPAPRR1604 pKa = 11.84SAPTPAPRR1612 pKa = 11.84APQAPSPTPAPTPAPRR1628 pKa = 11.84TAPTPAPRR1636 pKa = 11.84SAPTPAPRR1644 pKa = 11.84APQAPSPTPAPTPAPRR1660 pKa = 11.84TAPTPAPAPAPRR1672 pKa = 11.84SAPTPAPRR1680 pKa = 11.84APQAPSPTPAPTPAPMTAPTPSPRR1704 pKa = 11.84SAPTPAPRR1712 pKa = 11.84APQAPSPTPAPTPAPRR1728 pKa = 11.84TAPTPAPRR1736 pKa = 11.84SAPTPAPRR1744 pKa = 11.84APQAPSPTPAPTPTPRR1760 pKa = 11.84TAPTPAPAPAPRR1772 pKa = 11.84SAPTPAPRR1780 pKa = 11.84APQAPSPTPAPTPAPMTAPTPAPAPAPRR1808 pKa = 11.84SAPTPAPRR1816 pKa = 11.84APQAPSPTPPPTPAPRR1832 pKa = 11.84TAPTLAPTPAPRR1844 pKa = 11.84SAPTPAPRR1852 pKa = 11.84APLVPSPTPAPTPAPRR1868 pKa = 11.84MAPTPAPRR1876 pKa = 11.84SAPTPAPRR1884 pKa = 11.84APQAPSPTRR1893 pKa = 11.84APTPAPRR1900 pKa = 11.84TAPTPAPIAAPRR1912 pKa = 11.84SAPTPAPRR1920 pKa = 11.84APQAPSPTPAPTPVPMLAPTPVPAPTPTPARR1951 pKa = 11.84TPAPTPAPTPTPTSAPTPAPTPVPTPSPTPAPTSAPTPVPTPAPTPVPKK2000 pKa = 9.67PAPMPTLTPPPATPPPPPPPTPPPPPTPVPKK2031 pKa = 9.49PAPMPTLTPPPPTPPPPPPPTPPPPPPPTQSVCPAGFDD2069 pKa = 3.48DD2070 pKa = 5.65SKK2072 pKa = 11.63AFLSAIPVYY2081 pKa = 10.57LKK2083 pKa = 10.91RR2084 pKa = 11.84NILLLIVSKK2093 pKa = 10.93NIADD2097 pKa = 4.24ASPGTSTTKK2106 pKa = 10.38CLCYY2110 pKa = 10.59YY2111 pKa = 8.77STNLYY2116 pKa = 10.66LPIPTPNVKK2125 pKa = 10.41GPIACRR2131 pKa = 11.84RR2132 pKa = 11.84GG2133 pKa = 3.21
MM1 pKa = 7.85ASRR4 pKa = 11.84SITYY8 pKa = 9.4PGLLLLVVALVIVPSSQAKK27 pKa = 9.64RR28 pKa = 11.84SPPRR32 pKa = 11.84SAPTPAPRR40 pKa = 11.84TAPSPAPRR48 pKa = 11.84SAPTPAPRR56 pKa = 11.84SAPTPAPRR64 pKa = 11.84APQAPSPTPAPTPAPRR80 pKa = 11.84TAPTPAPRR88 pKa = 11.84SAPTPAPRR96 pKa = 11.84APQAPSPTPAPKK108 pKa = 9.98PPPRR112 pKa = 11.84TAPTTAPTPAPRR124 pKa = 11.84SAPTPAPRR132 pKa = 11.84APQAPSPTPAPTPAPTPAPRR152 pKa = 11.84TAPTPAPRR160 pKa = 11.84SAPTPAPRR168 pKa = 11.84APQAPSPTPAPMPAPRR184 pKa = 11.84TAPTLAPRR192 pKa = 11.84SAPTPAPRR200 pKa = 11.84APQAPSPTPAPTPAPRR216 pKa = 11.84TAPISAPTPAPRR228 pKa = 11.84SAPTPTPRR236 pKa = 11.84APQAPSPTPAPTPAPRR252 pKa = 11.84TAPTQAPTPAPRR264 pKa = 11.84SAPTPAPRR272 pKa = 11.84APQVPSPTPAPTPAPRR288 pKa = 11.84TAPTPAPTPAPRR300 pKa = 11.84STPTPAPRR308 pKa = 11.84APQAPSPTPAPTPAPRR324 pKa = 11.84TAPTPAPTPTPRR336 pKa = 11.84SAPTPAPRR344 pKa = 11.84APQVPSPTPTPTPAPRR360 pKa = 11.84TAPTPAPRR368 pKa = 11.84SAPTPAPRR376 pKa = 11.84APQAPSPTPAPTPAPRR392 pKa = 11.84TAPTPAPTPAPRR404 pKa = 11.84SAPTPAPRR412 pKa = 11.84APQAPSPTPTPTPAPRR428 pKa = 11.84TAPTPAPTPAPTPAPRR444 pKa = 11.84SAPTPAPRR452 pKa = 11.84APQAPSPTPAPTPAPRR468 pKa = 11.84TAPTPAPRR476 pKa = 11.84SAPTPAPRR484 pKa = 11.84APQAPSPTPTPTPAPRR500 pKa = 11.84TAPTPAPTPAPTPAPRR516 pKa = 11.84SAPTPAPRR524 pKa = 11.84APQAPSPTPAPTPAPRR540 pKa = 11.84TAPTPAPRR548 pKa = 11.84SAPTPAPRR556 pKa = 11.84APQAPSPTPAPTPAPRR572 pKa = 11.84TAPTPAPRR580 pKa = 11.84SAPTPAPRR588 pKa = 11.84APQAPSPTPAPTPAPRR604 pKa = 11.84TAPTPAPRR612 pKa = 11.84SAPTPAPRR620 pKa = 11.84APQAPSPTPAPTPAPRR636 pKa = 11.84MAPTPASTPAPRR648 pKa = 11.84SAPTPAPRR656 pKa = 11.84ASQAPSPTPAPTPAPRR672 pKa = 11.84MAPTPAPRR680 pKa = 11.84SVPTPAPRR688 pKa = 11.84APLAPSPTPAPTLAPRR704 pKa = 11.84TAPTPAPRR712 pKa = 11.84SAPTLAPRR720 pKa = 11.84APQAPSPRR728 pKa = 11.84PAPTPAPRR736 pKa = 11.84TAPTPAPTPTPRR748 pKa = 11.84SAPSPAPRR756 pKa = 11.84APQVPSPTRR765 pKa = 11.84APTPAPRR772 pKa = 11.84TAPTMAPTPAPRR784 pKa = 11.84SAPTPAPRR792 pKa = 11.84APQAPSPTPAPTPAPRR808 pKa = 11.84TAPTPAPTPAPRR820 pKa = 11.84SAPTPAPRR828 pKa = 11.84APQAPSPTHH837 pKa = 6.79APMLAPRR844 pKa = 11.84TAPTPAPTPTPRR856 pKa = 11.84LAPTPAPRR864 pKa = 11.84APQAPSPTTTPTPAPRR880 pKa = 11.84TAPTPTPRR888 pKa = 11.84SAPTPAPRR896 pKa = 11.84APQAPSPTPAPTQAPRR912 pKa = 11.84TAPTPAPTPAPRR924 pKa = 11.84SAPTPAPRR932 pKa = 11.84APQAPSPTPAPTPAPRR948 pKa = 11.84TAPTPAPTPAPRR960 pKa = 11.84SAPTPAPRR968 pKa = 11.84PPQAPSPTPAPTPASRR984 pKa = 11.84TAPTPSPRR992 pKa = 11.84SAPTPAPRR1000 pKa = 11.84APLAPSPTPAPTPAPRR1016 pKa = 11.84TAPTPAPRR1024 pKa = 11.84SAPTPAPRR1032 pKa = 11.84APQAPSPTPAPTPAPRR1048 pKa = 11.84TAPTPAPRR1056 pKa = 11.84SAPTPTPRR1064 pKa = 11.84APQAPSPTPAPTPAPRR1080 pKa = 11.84MAPTPAPTPAPRR1092 pKa = 11.84SAPTPAPRR1100 pKa = 11.84ASQAPSPTPAPTPAPRR1116 pKa = 11.84MAPTPAPRR1124 pKa = 11.84SVPTPAPRR1132 pKa = 11.84APLAPSPTPAPTLAPRR1148 pKa = 11.84TAPTPAPRR1156 pKa = 11.84SAPTLAPRR1164 pKa = 11.84APQAPSPTPAPTPAPRR1180 pKa = 11.84TAPTPAPTPTPRR1192 pKa = 11.84SAPSPAPRR1200 pKa = 11.84APQVPSPTPAPTPAPRR1216 pKa = 11.84TAPTTAPTPAPRR1228 pKa = 11.84SAPTPAPRR1236 pKa = 11.84APQAPSPTPAPTPAPRR1252 pKa = 11.84TAPTPAPTPTPRR1264 pKa = 11.84SAPTPAPRR1272 pKa = 11.84APQAPSPTPAPMLAPRR1288 pKa = 11.84TAPTPAPTPTPRR1300 pKa = 11.84LAPTPAPRR1308 pKa = 11.84APQAPSPTPTPTPAPRR1324 pKa = 11.84MAPTPAPRR1332 pKa = 11.84SAPTPAPRR1340 pKa = 11.84APQAPSPTPAPTQAPRR1356 pKa = 11.84TAPTPAPTPAPRR1368 pKa = 11.84SAPTPAPRR1376 pKa = 11.84APQAPSPTPAPTPAPRR1392 pKa = 11.84TAPTPAPTPAPRR1404 pKa = 11.84SAPTPTPRR1412 pKa = 11.84PPQAPSPTPAPTPASRR1428 pKa = 11.84TAPTPAPRR1436 pKa = 11.84SAPTPAPRR1444 pKa = 11.84APQAPSPTPAPRR1456 pKa = 11.84TAPTPAPRR1464 pKa = 11.84SAPTPAPRR1472 pKa = 11.84APQAPSPTPAPTPAPRR1488 pKa = 11.84TAPTPAPTPAPRR1500 pKa = 11.84FAPTPAPRR1508 pKa = 11.84APQAPSPTPAPTPAPRR1524 pKa = 11.84TAPTPAPTPAPRR1536 pKa = 11.84SAPTLAPRR1544 pKa = 11.84APQAPSPTPTPTPAPRR1560 pKa = 11.84TAPTPAPTPAPRR1572 pKa = 11.84SAPTPAPRR1580 pKa = 11.84APQAPSPTPAPTPAPRR1596 pKa = 11.84TAPTPAPRR1604 pKa = 11.84SAPTPAPRR1612 pKa = 11.84APQAPSPTPAPTPAPRR1628 pKa = 11.84TAPTPAPRR1636 pKa = 11.84SAPTPAPRR1644 pKa = 11.84APQAPSPTPAPTPAPRR1660 pKa = 11.84TAPTPAPAPAPRR1672 pKa = 11.84SAPTPAPRR1680 pKa = 11.84APQAPSPTPAPTPAPMTAPTPSPRR1704 pKa = 11.84SAPTPAPRR1712 pKa = 11.84APQAPSPTPAPTPAPRR1728 pKa = 11.84TAPTPAPRR1736 pKa = 11.84SAPTPAPRR1744 pKa = 11.84APQAPSPTPAPTPTPRR1760 pKa = 11.84TAPTPAPAPAPRR1772 pKa = 11.84SAPTPAPRR1780 pKa = 11.84APQAPSPTPAPTPAPMTAPTPAPAPAPRR1808 pKa = 11.84SAPTPAPRR1816 pKa = 11.84APQAPSPTPPPTPAPRR1832 pKa = 11.84TAPTLAPTPAPRR1844 pKa = 11.84SAPTPAPRR1852 pKa = 11.84APLVPSPTPAPTPAPRR1868 pKa = 11.84MAPTPAPRR1876 pKa = 11.84SAPTPAPRR1884 pKa = 11.84APQAPSPTRR1893 pKa = 11.84APTPAPRR1900 pKa = 11.84TAPTPAPIAAPRR1912 pKa = 11.84SAPTPAPRR1920 pKa = 11.84APQAPSPTPAPTPVPMLAPTPVPAPTPTPARR1951 pKa = 11.84TPAPTPAPTPTPTSAPTPAPTPVPTPSPTPAPTSAPTPVPTPAPTPVPKK2000 pKa = 9.67PAPMPTLTPPPATPPPPPPPTPPPPPTPVPKK2031 pKa = 9.49PAPMPTLTPPPPTPPPPPPPTPPPPPPPTQSVCPAGFDD2069 pKa = 3.48DD2070 pKa = 5.65SKK2072 pKa = 11.63AFLSAIPVYY2081 pKa = 10.57LKK2083 pKa = 10.91RR2084 pKa = 11.84NILLLIVSKK2093 pKa = 10.93NIADD2097 pKa = 4.24ASPGTSTTKK2106 pKa = 10.38CLCYY2110 pKa = 10.59YY2111 pKa = 8.77STNLYY2116 pKa = 10.66LPIPTPNVKK2125 pKa = 10.41GPIACRR2131 pKa = 11.84RR2132 pKa = 11.84GG2133 pKa = 3.21
Molecular weight: 208.95 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
18246505 |
29 |
5148 |
392.3 |
43.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.183 ± 0.017 | 1.974 ± 0.006 |
5.337 ± 0.008 | 5.994 ± 0.014 |
3.655 ± 0.007 | 7.252 ± 0.013 |
2.498 ± 0.006 | 4.383 ± 0.009 |
4.926 ± 0.011 | 9.583 ± 0.016 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.407 ± 0.004 | 3.524 ± 0.009 |
5.727 ± 0.015 | 3.541 ± 0.01 |
6.337 ± 0.011 | 8.39 ± 0.013 |
4.768 ± 0.007 | 6.659 ± 0.009 |
1.315 ± 0.004 | 2.547 ± 0.006 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
