
Mycobacterium virus Babsiella
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; Chebruvirinae; Brujitavirus
Average proteome isoelectric point is 6.13
Get precalculated fractions of proteins
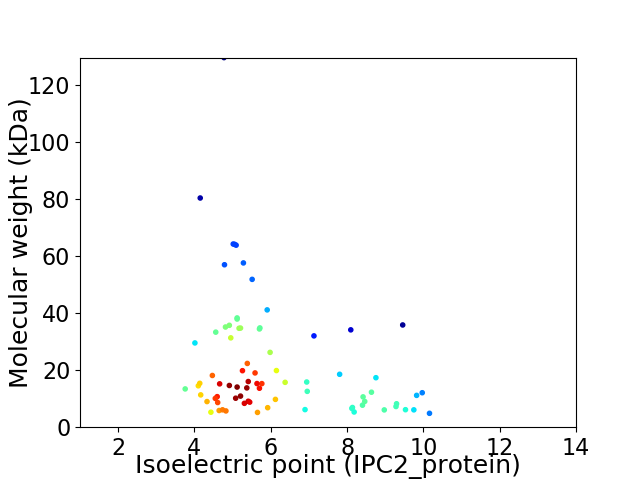
Virtual 2D-PAGE plot for 78 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|G8I6Q7|G8I6Q7_9CAUD Uncharacterized protein OS=Mycobacterium virus Babsiella OX=1089115 GN=24 PE=4 SV=1
MM1 pKa = 7.49EE2 pKa = 5.87LFVPCPLCGSDD13 pKa = 3.44VAVPLLSEE21 pKa = 4.46GDD23 pKa = 3.7HH24 pKa = 6.42VRR26 pKa = 11.84APDD29 pKa = 4.25LLPNMNSHH37 pKa = 6.92VISVHH42 pKa = 5.19EE43 pKa = 4.08MDD45 pKa = 3.8VSGMM49 pKa = 3.65
MM1 pKa = 7.49EE2 pKa = 5.87LFVPCPLCGSDD13 pKa = 3.44VAVPLLSEE21 pKa = 4.46GDD23 pKa = 3.7HH24 pKa = 6.42VRR26 pKa = 11.84APDD29 pKa = 4.25LLPNMNSHH37 pKa = 6.92VISVHH42 pKa = 5.19EE43 pKa = 4.08MDD45 pKa = 3.8VSGMM49 pKa = 3.65
Molecular weight: 5.26 kDa
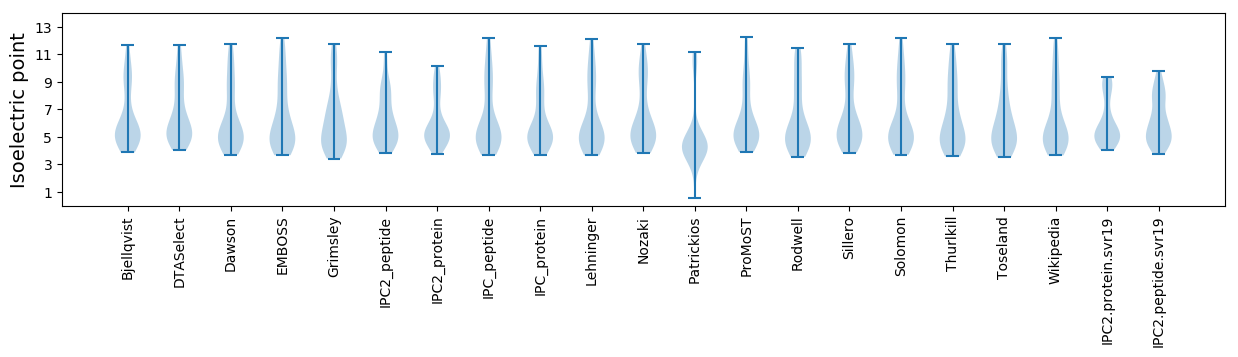
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|G8I6R7|G8I6R7_9CAUD Uncharacterized protein OS=Mycobacterium virus Babsiella OX=1089115 GN=34 PE=4 SV=1
MM1 pKa = 7.82LLEE4 pKa = 4.52QKK6 pKa = 10.65FSPQSVGSVSRR17 pKa = 11.84VHH19 pKa = 7.21GPMVNRR25 pKa = 11.84WRR27 pKa = 11.84TWQFAQSLSRR37 pKa = 11.84RR38 pKa = 11.84TVEE41 pKa = 3.71EE42 pKa = 3.55RR43 pKa = 11.84VATVRR48 pKa = 11.84RR49 pKa = 11.84MAEE52 pKa = 3.56WCGVSPEE59 pKa = 4.09FAQVEE64 pKa = 4.5QIVEE68 pKa = 3.97WLAEE72 pKa = 4.18GGTWSARR79 pKa = 11.84TRR81 pKa = 11.84WTYY84 pKa = 11.34YY85 pKa = 10.31GALSAWFLWLQQQGYY100 pKa = 8.47RR101 pKa = 11.84QDD103 pKa = 3.5NPMVMIGRR111 pKa = 11.84PKK113 pKa = 10.09RR114 pKa = 11.84PKK116 pKa = 9.75SVPRR120 pKa = 11.84PVSNLDD126 pKa = 3.3MQRR129 pKa = 11.84LLAVRR134 pKa = 11.84AHH136 pKa = 6.47KK137 pKa = 9.25RR138 pKa = 11.84TKK140 pKa = 11.33AMILLAAFQGLRR152 pKa = 11.84VHH154 pKa = 7.42EE155 pKa = 4.17IAQVKK160 pKa = 10.12GEE162 pKa = 4.11HH163 pKa = 6.82LDD165 pKa = 4.08LIEE168 pKa = 4.13RR169 pKa = 11.84TMTVTGKK176 pKa = 11.18GNITATLPLHH186 pKa = 5.86HH187 pKa = 6.81RR188 pKa = 11.84VVEE191 pKa = 4.17MAYY194 pKa = 9.4QMPRR198 pKa = 11.84KK199 pKa = 9.17GHH201 pKa = 6.4WFPGPDD207 pKa = 2.81RR208 pKa = 11.84GHH210 pKa = 5.51QRR212 pKa = 11.84RR213 pKa = 11.84EE214 pKa = 4.31SVSGTIKK221 pKa = 10.28EE222 pKa = 4.01AMIRR226 pKa = 11.84AGVLGSAHH234 pKa = 7.05CLRR237 pKa = 11.84HH238 pKa = 4.94WFGTALLEE246 pKa = 4.69AGVDD250 pKa = 3.56LRR252 pKa = 11.84TVQEE256 pKa = 4.11LMRR259 pKa = 11.84HH260 pKa = 4.7QSLTSTEE267 pKa = 3.62IYY269 pKa = 10.3TRR271 pKa = 11.84VTDD274 pKa = 3.31QRR276 pKa = 11.84RR277 pKa = 11.84AEE279 pKa = 4.72GIQRR283 pKa = 11.84LDD285 pKa = 3.27PFGVARR291 pKa = 11.84QGHH294 pKa = 5.86KK295 pKa = 10.41AADD298 pKa = 3.78IIGEE302 pKa = 4.19MIADD306 pKa = 4.18EE307 pKa = 4.42VAGSGASSAAA317 pKa = 3.37
MM1 pKa = 7.82LLEE4 pKa = 4.52QKK6 pKa = 10.65FSPQSVGSVSRR17 pKa = 11.84VHH19 pKa = 7.21GPMVNRR25 pKa = 11.84WRR27 pKa = 11.84TWQFAQSLSRR37 pKa = 11.84RR38 pKa = 11.84TVEE41 pKa = 3.71EE42 pKa = 3.55RR43 pKa = 11.84VATVRR48 pKa = 11.84RR49 pKa = 11.84MAEE52 pKa = 3.56WCGVSPEE59 pKa = 4.09FAQVEE64 pKa = 4.5QIVEE68 pKa = 3.97WLAEE72 pKa = 4.18GGTWSARR79 pKa = 11.84TRR81 pKa = 11.84WTYY84 pKa = 11.34YY85 pKa = 10.31GALSAWFLWLQQQGYY100 pKa = 8.47RR101 pKa = 11.84QDD103 pKa = 3.5NPMVMIGRR111 pKa = 11.84PKK113 pKa = 10.09RR114 pKa = 11.84PKK116 pKa = 9.75SVPRR120 pKa = 11.84PVSNLDD126 pKa = 3.3MQRR129 pKa = 11.84LLAVRR134 pKa = 11.84AHH136 pKa = 6.47KK137 pKa = 9.25RR138 pKa = 11.84TKK140 pKa = 11.33AMILLAAFQGLRR152 pKa = 11.84VHH154 pKa = 7.42EE155 pKa = 4.17IAQVKK160 pKa = 10.12GEE162 pKa = 4.11HH163 pKa = 6.82LDD165 pKa = 4.08LIEE168 pKa = 4.13RR169 pKa = 11.84TMTVTGKK176 pKa = 11.18GNITATLPLHH186 pKa = 5.86HH187 pKa = 6.81RR188 pKa = 11.84VVEE191 pKa = 4.17MAYY194 pKa = 9.4QMPRR198 pKa = 11.84KK199 pKa = 9.17GHH201 pKa = 6.4WFPGPDD207 pKa = 2.81RR208 pKa = 11.84GHH210 pKa = 5.51QRR212 pKa = 11.84RR213 pKa = 11.84EE214 pKa = 4.31SVSGTIKK221 pKa = 10.28EE222 pKa = 4.01AMIRR226 pKa = 11.84AGVLGSAHH234 pKa = 7.05CLRR237 pKa = 11.84HH238 pKa = 4.94WFGTALLEE246 pKa = 4.69AGVDD250 pKa = 3.56LRR252 pKa = 11.84TVQEE256 pKa = 4.11LMRR259 pKa = 11.84HH260 pKa = 4.7QSLTSTEE267 pKa = 3.62IYY269 pKa = 10.3TRR271 pKa = 11.84VTDD274 pKa = 3.31QRR276 pKa = 11.84RR277 pKa = 11.84AEE279 pKa = 4.72GIQRR283 pKa = 11.84LDD285 pKa = 3.27PFGVARR291 pKa = 11.84QGHH294 pKa = 5.86KK295 pKa = 10.41AADD298 pKa = 3.78IIGEE302 pKa = 4.19MIADD306 pKa = 4.18EE307 pKa = 4.42VAGSGASSAAA317 pKa = 3.37
Molecular weight: 35.84 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
15845 |
40 |
1255 |
203.1 |
22.13 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.562 ± 0.499 | 0.947 ± 0.129 |
6.854 ± 0.244 | 6.015 ± 0.275 |
2.979 ± 0.184 | 9.183 ± 0.554 |
2.057 ± 0.176 | 4.361 ± 0.167 |
3.238 ± 0.22 | 7.889 ± 0.296 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.152 ± 0.133 | 3.08 ± 0.192 |
6.191 ± 0.202 | 3.812 ± 0.247 |
7.176 ± 0.492 | 5.308 ± 0.262 |
5.92 ± 0.298 | 6.835 ± 0.248 |
2.045 ± 0.122 | 2.398 ± 0.158 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
