
Human papillomavirus 120
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Betapapillomavirus; Betapapillomavirus 2
Average proteome isoelectric point is 5.89
Get precalculated fractions of proteins
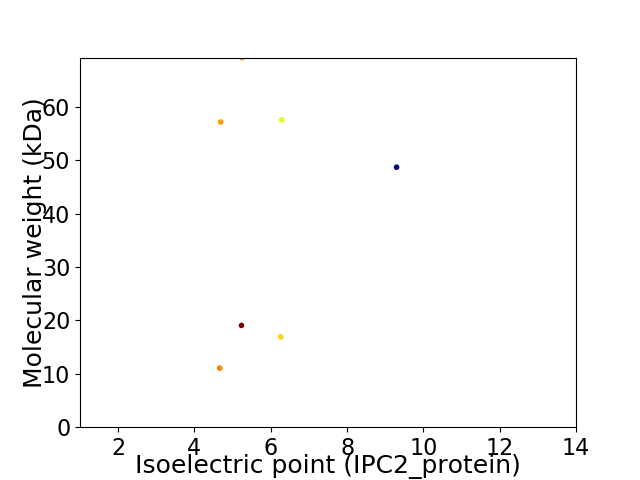
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|D7P169|D7P169_9PAPI Regulatory protein E2 OS=Human papillomavirus 120 OX=765052 GN=E2 PE=3 SV=1
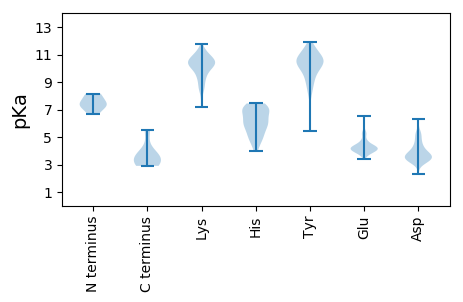
MM1 pKa = 7.54IGKK4 pKa = 8.59QATLRR9 pKa = 11.84DD10 pKa = 3.69IVLEE14 pKa = 4.01EE15 pKa = 4.08LVQPIDD21 pKa = 3.57LHH23 pKa = 6.18CHH25 pKa = 5.48EE26 pKa = 5.19EE27 pKa = 3.89LLEE30 pKa = 4.19EE31 pKa = 4.18VEE33 pKa = 4.19EE34 pKa = 4.2AVVEE38 pKa = 4.18EE39 pKa = 4.3EE40 pKa = 5.26PEE42 pKa = 3.92YY43 pKa = 10.56TPYY46 pKa = 10.77KK47 pKa = 10.31IIVNCGGCDD56 pKa = 3.18TKK58 pKa = 11.4LKK60 pKa = 10.72LFVLATDD67 pKa = 3.87FGIRR71 pKa = 11.84AFQASLLEE79 pKa = 4.13NVRR82 pKa = 11.84LVCPSCRR89 pKa = 11.84EE90 pKa = 4.15DD91 pKa = 3.16IRR93 pKa = 11.84DD94 pKa = 3.4GRR96 pKa = 11.84RR97 pKa = 2.97
MM1 pKa = 7.54IGKK4 pKa = 8.59QATLRR9 pKa = 11.84DD10 pKa = 3.69IVLEE14 pKa = 4.01EE15 pKa = 4.08LVQPIDD21 pKa = 3.57LHH23 pKa = 6.18CHH25 pKa = 5.48EE26 pKa = 5.19EE27 pKa = 3.89LLEE30 pKa = 4.19EE31 pKa = 4.18VEE33 pKa = 4.19EE34 pKa = 4.2AVVEE38 pKa = 4.18EE39 pKa = 4.3EE40 pKa = 5.26PEE42 pKa = 3.92YY43 pKa = 10.56TPYY46 pKa = 10.77KK47 pKa = 10.31IIVNCGGCDD56 pKa = 3.18TKK58 pKa = 11.4LKK60 pKa = 10.72LFVLATDD67 pKa = 3.87FGIRR71 pKa = 11.84AFQASLLEE79 pKa = 4.13NVRR82 pKa = 11.84LVCPSCRR89 pKa = 11.84EE90 pKa = 4.15DD91 pKa = 3.16IRR93 pKa = 11.84DD94 pKa = 3.4GRR96 pKa = 11.84RR97 pKa = 2.97
Molecular weight: 11.07 kDa
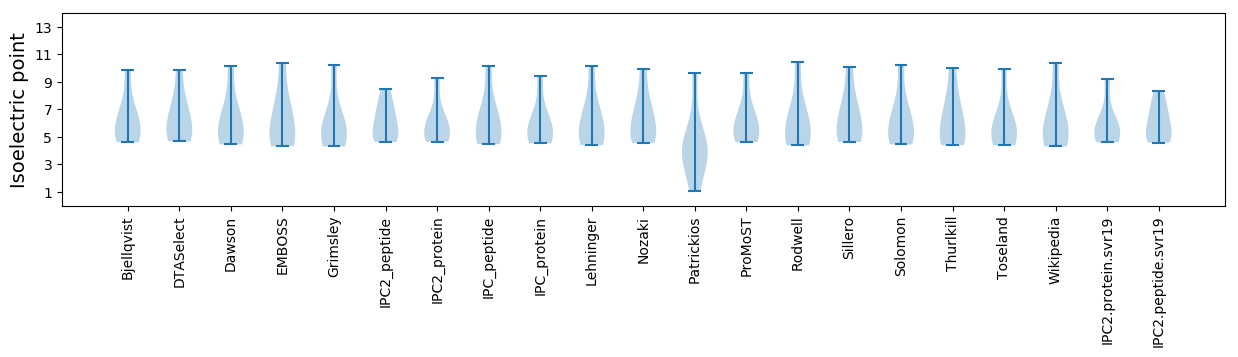
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D7P170|D7P170_9PAPI E4 OS=Human papillomavirus 120 OX=765052 GN=E4 PE=4 SV=1
MM1 pKa = 7.43EE2 pKa = 5.41ALSEE6 pKa = 4.17RR7 pKa = 11.84FSALQEE13 pKa = 4.13KK14 pKa = 10.62LMDD17 pKa = 4.39LYY19 pKa = 11.33EE20 pKa = 5.54SGLEE24 pKa = 3.93DD25 pKa = 5.27LEE27 pKa = 4.38TQILHH32 pKa = 6.0WKK34 pKa = 9.93LLRR37 pKa = 11.84QEE39 pKa = 4.47QIIFYY44 pKa = 8.08YY45 pKa = 10.09AKK47 pKa = 10.23KK48 pKa = 10.42RR49 pKa = 11.84GITRR53 pKa = 11.84LGYY56 pKa = 10.29QPVPSLASSEE66 pKa = 4.31TKK68 pKa = 10.71AKK70 pKa = 10.48DD71 pKa = 3.41AIAIGLLLEE80 pKa = 4.52SLQKK84 pKa = 10.35SQYY87 pKa = 11.21ANEE90 pKa = 3.95PWTLVEE96 pKa = 4.59TSLEE100 pKa = 4.5TIKK103 pKa = 10.95SPPVDD108 pKa = 3.9CFKK111 pKa = 10.9KK112 pKa = 10.17GPKK115 pKa = 8.36TVEE118 pKa = 4.01VYY120 pKa = 10.76FDD122 pKa = 3.87GNPANVMPYY131 pKa = 8.82TVWSYY136 pKa = 11.23IYY138 pKa = 10.22YY139 pKa = 8.39QTDD142 pKa = 2.95EE143 pKa = 4.38DD144 pKa = 3.6TWEE147 pKa = 4.14KK148 pKa = 11.38VEE150 pKa = 4.12GQVDD154 pKa = 3.59YY155 pKa = 10.8TGAYY159 pKa = 9.48FFEE162 pKa = 5.24GNFKK166 pKa = 10.59NYY168 pKa = 9.99YY169 pKa = 9.78IKK171 pKa = 10.8FEE173 pKa = 4.37NDD175 pKa = 2.48AKK177 pKa = 10.83RR178 pKa = 11.84YY179 pKa = 5.43GTTGIWEE186 pKa = 4.09VHH188 pKa = 4.93VDD190 pKa = 3.26KK191 pKa = 10.63DD192 pKa = 4.18TVFTPVTSSTPPVGDD207 pKa = 3.52ASNNTVPEE215 pKa = 4.14AVSASVSSPQRR226 pKa = 11.84SPSTNRR232 pKa = 11.84RR233 pKa = 11.84YY234 pKa = 9.89GRR236 pKa = 11.84KK237 pKa = 9.3ASSPTTTSRR246 pKa = 11.84RR247 pKa = 11.84QKK249 pKa = 9.81RR250 pKa = 11.84QRR252 pKa = 11.84EE253 pKa = 3.87EE254 pKa = 3.38TRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84KK259 pKa = 7.63TRR261 pKa = 11.84SRR263 pKa = 11.84SRR265 pKa = 11.84SRR267 pKa = 11.84EE268 pKa = 3.58QRR270 pKa = 11.84GGRR273 pKa = 11.84EE274 pKa = 3.96TRR276 pKa = 11.84RR277 pKa = 11.84SSSRR281 pKa = 11.84GSSKK285 pKa = 10.51SPRR288 pKa = 11.84RR289 pKa = 11.84GGNGGGGPLTRR300 pKa = 11.84SRR302 pKa = 11.84SRR304 pKa = 11.84SRR306 pKa = 11.84TRR308 pKa = 11.84KK309 pKa = 8.73EE310 pKa = 3.89SVGGGGVAPSEE321 pKa = 4.16VGATVRR327 pKa = 11.84SLSRR331 pKa = 11.84HH332 pKa = 4.35NSGRR336 pKa = 11.84LAQLLDD342 pKa = 3.62AAKK345 pKa = 9.99DD346 pKa = 3.57PPVILLRR353 pKa = 11.84GNANTLKK360 pKa = 10.22CYY362 pKa = 9.81RR363 pKa = 11.84WRR365 pKa = 11.84FRR367 pKa = 11.84KK368 pKa = 9.42KK369 pKa = 8.94HH370 pKa = 5.52AGNFRR375 pKa = 11.84FVSTTWSWVGGNSNDD390 pKa = 3.73RR391 pKa = 11.84IGRR394 pKa = 11.84ARR396 pKa = 11.84MLIAFDD402 pKa = 3.85TDD404 pKa = 3.78LEE406 pKa = 4.21RR407 pKa = 11.84DD408 pKa = 3.28KK409 pKa = 11.25CIQQMKK415 pKa = 10.18LPLGVDD421 pKa = 3.33WSYY424 pKa = 11.93GQFDD428 pKa = 4.23DD429 pKa = 5.18LL430 pKa = 5.54
MM1 pKa = 7.43EE2 pKa = 5.41ALSEE6 pKa = 4.17RR7 pKa = 11.84FSALQEE13 pKa = 4.13KK14 pKa = 10.62LMDD17 pKa = 4.39LYY19 pKa = 11.33EE20 pKa = 5.54SGLEE24 pKa = 3.93DD25 pKa = 5.27LEE27 pKa = 4.38TQILHH32 pKa = 6.0WKK34 pKa = 9.93LLRR37 pKa = 11.84QEE39 pKa = 4.47QIIFYY44 pKa = 8.08YY45 pKa = 10.09AKK47 pKa = 10.23KK48 pKa = 10.42RR49 pKa = 11.84GITRR53 pKa = 11.84LGYY56 pKa = 10.29QPVPSLASSEE66 pKa = 4.31TKK68 pKa = 10.71AKK70 pKa = 10.48DD71 pKa = 3.41AIAIGLLLEE80 pKa = 4.52SLQKK84 pKa = 10.35SQYY87 pKa = 11.21ANEE90 pKa = 3.95PWTLVEE96 pKa = 4.59TSLEE100 pKa = 4.5TIKK103 pKa = 10.95SPPVDD108 pKa = 3.9CFKK111 pKa = 10.9KK112 pKa = 10.17GPKK115 pKa = 8.36TVEE118 pKa = 4.01VYY120 pKa = 10.76FDD122 pKa = 3.87GNPANVMPYY131 pKa = 8.82TVWSYY136 pKa = 11.23IYY138 pKa = 10.22YY139 pKa = 8.39QTDD142 pKa = 2.95EE143 pKa = 4.38DD144 pKa = 3.6TWEE147 pKa = 4.14KK148 pKa = 11.38VEE150 pKa = 4.12GQVDD154 pKa = 3.59YY155 pKa = 10.8TGAYY159 pKa = 9.48FFEE162 pKa = 5.24GNFKK166 pKa = 10.59NYY168 pKa = 9.99YY169 pKa = 9.78IKK171 pKa = 10.8FEE173 pKa = 4.37NDD175 pKa = 2.48AKK177 pKa = 10.83RR178 pKa = 11.84YY179 pKa = 5.43GTTGIWEE186 pKa = 4.09VHH188 pKa = 4.93VDD190 pKa = 3.26KK191 pKa = 10.63DD192 pKa = 4.18TVFTPVTSSTPPVGDD207 pKa = 3.52ASNNTVPEE215 pKa = 4.14AVSASVSSPQRR226 pKa = 11.84SPSTNRR232 pKa = 11.84RR233 pKa = 11.84YY234 pKa = 9.89GRR236 pKa = 11.84KK237 pKa = 9.3ASSPTTTSRR246 pKa = 11.84RR247 pKa = 11.84QKK249 pKa = 9.81RR250 pKa = 11.84QRR252 pKa = 11.84EE253 pKa = 3.87EE254 pKa = 3.38TRR256 pKa = 11.84RR257 pKa = 11.84RR258 pKa = 11.84KK259 pKa = 7.63TRR261 pKa = 11.84SRR263 pKa = 11.84SRR265 pKa = 11.84SRR267 pKa = 11.84EE268 pKa = 3.58QRR270 pKa = 11.84GGRR273 pKa = 11.84EE274 pKa = 3.96TRR276 pKa = 11.84RR277 pKa = 11.84SSSRR281 pKa = 11.84GSSKK285 pKa = 10.51SPRR288 pKa = 11.84RR289 pKa = 11.84GGNGGGGPLTRR300 pKa = 11.84SRR302 pKa = 11.84SRR304 pKa = 11.84SRR306 pKa = 11.84TRR308 pKa = 11.84KK309 pKa = 8.73EE310 pKa = 3.89SVGGGGVAPSEE321 pKa = 4.16VGATVRR327 pKa = 11.84SLSRR331 pKa = 11.84HH332 pKa = 4.35NSGRR336 pKa = 11.84LAQLLDD342 pKa = 3.62AAKK345 pKa = 9.99DD346 pKa = 3.57PPVILLRR353 pKa = 11.84GNANTLKK360 pKa = 10.22CYY362 pKa = 9.81RR363 pKa = 11.84WRR365 pKa = 11.84FRR367 pKa = 11.84KK368 pKa = 9.42KK369 pKa = 8.94HH370 pKa = 5.52AGNFRR375 pKa = 11.84FVSTTWSWVGGNSNDD390 pKa = 3.73RR391 pKa = 11.84IGRR394 pKa = 11.84ARR396 pKa = 11.84MLIAFDD402 pKa = 3.85TDD404 pKa = 3.78LEE406 pKa = 4.21RR407 pKa = 11.84DD408 pKa = 3.28KK409 pKa = 11.25CIQQMKK415 pKa = 10.18LPLGVDD421 pKa = 3.33WSYY424 pKa = 11.93GQFDD428 pKa = 4.23DD429 pKa = 5.18LL430 pKa = 5.54
Molecular weight: 48.71 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2472 |
97 |
604 |
353.1 |
39.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.583 ± 0.414 | 2.144 ± 0.719 |
6.472 ± 0.443 | 6.877 ± 0.719 |
4.207 ± 0.305 | 6.23 ± 0.608 |
1.739 ± 0.191 | 5.299 ± 0.883 |
5.421 ± 0.87 | 8.455 ± 0.649 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.578 ± 0.375 | 4.086 ± 0.506 |
6.553 ± 1.155 | 5.016 ± 0.67 |
6.432 ± 1.019 | 6.998 ± 0.84 |
5.785 ± 0.942 | 6.23 ± 0.72 |
1.294 ± 0.355 | 3.6 ± 0.427 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
