
Plasmodium falciparum FCH/4
Taxonomy: cellular organisms; Eukaryota; Sar; Alveolata; Apicomplexa; Aconoidasida; Haemosporida; Plasmodiidae; Plasmodium; Plasmodium (Laverania); Plasmodium falciparum
Average proteome isoelectric point is 7.21
Get precalculated fractions of proteins
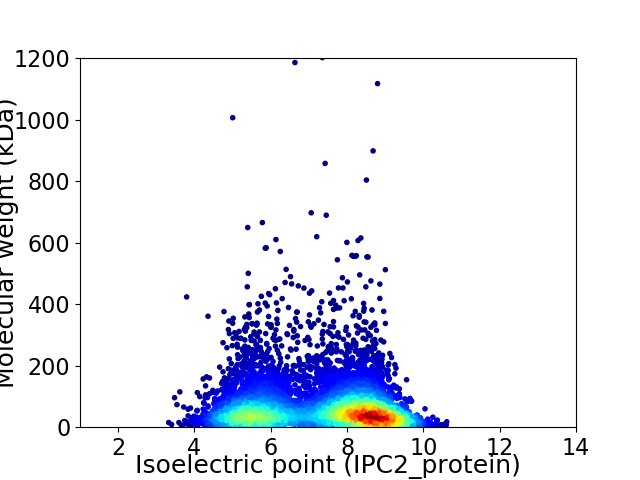
Virtual 2D-PAGE plot for 5762 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A024VR15|A0A024VR15_PLAFA Uncharacterized protein OS=Plasmodium falciparum FCH/4 OX=1036724 GN=PFFCH_01788 PE=4 SV=1
MM1 pKa = 8.65LEE3 pKa = 3.94EE4 pKa = 4.27RR5 pKa = 11.84NEE7 pKa = 3.97NNEE10 pKa = 3.93NNEE13 pKa = 4.01DD14 pKa = 3.92NEE16 pKa = 4.49NNGYY20 pKa = 8.4NQNNEE25 pKa = 3.51YY26 pKa = 10.56DD27 pKa = 3.53QNNEE31 pKa = 3.35YY32 pKa = 10.64DD33 pKa = 3.51QNNEE37 pKa = 3.35YY38 pKa = 10.64DD39 pKa = 3.51QNNEE43 pKa = 3.35YY44 pKa = 10.64DD45 pKa = 3.51QNNEE49 pKa = 3.35YY50 pKa = 10.64DD51 pKa = 3.51QNNEE55 pKa = 3.37YY56 pKa = 10.72DD57 pKa = 3.43QNNDD61 pKa = 3.01YY62 pKa = 10.88NQNNDD67 pKa = 3.17YY68 pKa = 10.78NQNNDD73 pKa = 3.17YY74 pKa = 10.78NQNNDD79 pKa = 3.17YY80 pKa = 10.78NQNNDD85 pKa = 3.17YY86 pKa = 10.78NQNNDD91 pKa = 3.06YY92 pKa = 10.96NLNSGYY98 pKa = 10.55NLNSGYY104 pKa = 10.36NLNSGYY110 pKa = 10.45SLNSGYY116 pKa = 10.98SLNNVYY122 pKa = 10.32NQYY125 pKa = 11.36SNNIEE130 pKa = 3.87NSGNILNNGFFGSSTNNNNRR150 pKa = 11.84SILGGVRR157 pKa = 11.84EE158 pKa = 4.58PIQLEE163 pKa = 4.0SRR165 pKa = 11.84GFFINGRR172 pKa = 11.84SNNVNRR178 pKa = 11.84ILVNVDD184 pKa = 3.25HH185 pKa = 7.1NNVPINSLNGTNHH198 pKa = 6.96ILGIPNNINMDD209 pKa = 4.0SLIQTILLASSLIINTSARR228 pKa = 11.84AVATSNRR235 pKa = 11.84NTRR238 pKa = 11.84NSNALTSDD246 pKa = 3.54GEE248 pKa = 4.28EE249 pKa = 4.05TMEE252 pKa = 4.89EE253 pKa = 4.03IVDD256 pKa = 4.02DD257 pKa = 4.22GMEE260 pKa = 4.32EE261 pKa = 3.7IDD263 pKa = 4.67LNEE266 pKa = 4.22NSDD269 pKa = 4.44FIHH272 pKa = 7.4DD273 pKa = 4.13GNPLGEE279 pKa = 4.28YY280 pKa = 10.96SNMEE284 pKa = 4.0YY285 pKa = 10.9EE286 pKa = 4.48LLNEE290 pKa = 4.48VSASTDD296 pKa = 3.38FSDD299 pKa = 3.5YY300 pKa = 11.01HH301 pKa = 7.75SVVDD305 pKa = 3.89EE306 pKa = 4.68DD307 pKa = 5.63LINFNDD313 pKa = 3.99DD314 pKa = 3.11VTLEE318 pKa = 4.2TQSMIAHH325 pKa = 6.92GGSLSEE331 pKa = 4.27IEE333 pKa = 4.17EE334 pKa = 4.39TGDD337 pKa = 3.66LSSDD341 pKa = 3.24VDD343 pKa = 3.74RR344 pKa = 11.84LLSSIEE350 pKa = 3.84TTPRR354 pKa = 11.84DD355 pKa = 3.48DD356 pKa = 3.26VVYY359 pKa = 10.56IRR361 pKa = 11.84GDD363 pKa = 3.33IILGEE368 pKa = 4.14DD369 pKa = 3.84EE370 pKa = 5.19IIIEE374 pKa = 4.18NGEE377 pKa = 4.12IILNNTDD384 pKa = 3.64EE385 pKa = 4.7LFEE388 pKa = 5.13DD389 pKa = 4.03VPEE392 pKa = 4.08MLEE395 pKa = 4.01NANLLEE401 pKa = 4.49DD402 pKa = 3.8RR403 pKa = 11.84EE404 pKa = 4.66HH405 pKa = 7.44VFEE408 pKa = 4.42YY409 pKa = 10.54NISVANEE416 pKa = 3.48NTTNQNSYY424 pKa = 10.71NFNAPQDD431 pKa = 4.02NTLTNYY437 pKa = 10.32NISRR441 pKa = 11.84FLQRR445 pKa = 11.84YY446 pKa = 6.69GTTTPNTQHH455 pKa = 6.75NISIHH460 pKa = 6.53RR461 pKa = 11.84DD462 pKa = 3.18GHH464 pKa = 5.2SLRR467 pKa = 11.84SDD469 pKa = 3.19TNIYY473 pKa = 10.86DD474 pKa = 4.61RR475 pKa = 11.84INGPSIRR482 pKa = 11.84EE483 pKa = 3.67NSLYY487 pKa = 10.28PGSFEE492 pKa = 3.77MAQYY496 pKa = 11.03ADD498 pKa = 4.72RR499 pKa = 11.84YY500 pKa = 10.15QMGHH504 pKa = 6.64TIAEE508 pKa = 4.43GFLYY512 pKa = 10.73
MM1 pKa = 8.65LEE3 pKa = 3.94EE4 pKa = 4.27RR5 pKa = 11.84NEE7 pKa = 3.97NNEE10 pKa = 3.93NNEE13 pKa = 4.01DD14 pKa = 3.92NEE16 pKa = 4.49NNGYY20 pKa = 8.4NQNNEE25 pKa = 3.51YY26 pKa = 10.56DD27 pKa = 3.53QNNEE31 pKa = 3.35YY32 pKa = 10.64DD33 pKa = 3.51QNNEE37 pKa = 3.35YY38 pKa = 10.64DD39 pKa = 3.51QNNEE43 pKa = 3.35YY44 pKa = 10.64DD45 pKa = 3.51QNNEE49 pKa = 3.35YY50 pKa = 10.64DD51 pKa = 3.51QNNEE55 pKa = 3.37YY56 pKa = 10.72DD57 pKa = 3.43QNNDD61 pKa = 3.01YY62 pKa = 10.88NQNNDD67 pKa = 3.17YY68 pKa = 10.78NQNNDD73 pKa = 3.17YY74 pKa = 10.78NQNNDD79 pKa = 3.17YY80 pKa = 10.78NQNNDD85 pKa = 3.17YY86 pKa = 10.78NQNNDD91 pKa = 3.06YY92 pKa = 10.96NLNSGYY98 pKa = 10.55NLNSGYY104 pKa = 10.36NLNSGYY110 pKa = 10.45SLNSGYY116 pKa = 10.98SLNNVYY122 pKa = 10.32NQYY125 pKa = 11.36SNNIEE130 pKa = 3.87NSGNILNNGFFGSSTNNNNRR150 pKa = 11.84SILGGVRR157 pKa = 11.84EE158 pKa = 4.58PIQLEE163 pKa = 4.0SRR165 pKa = 11.84GFFINGRR172 pKa = 11.84SNNVNRR178 pKa = 11.84ILVNVDD184 pKa = 3.25HH185 pKa = 7.1NNVPINSLNGTNHH198 pKa = 6.96ILGIPNNINMDD209 pKa = 4.0SLIQTILLASSLIINTSARR228 pKa = 11.84AVATSNRR235 pKa = 11.84NTRR238 pKa = 11.84NSNALTSDD246 pKa = 3.54GEE248 pKa = 4.28EE249 pKa = 4.05TMEE252 pKa = 4.89EE253 pKa = 4.03IVDD256 pKa = 4.02DD257 pKa = 4.22GMEE260 pKa = 4.32EE261 pKa = 3.7IDD263 pKa = 4.67LNEE266 pKa = 4.22NSDD269 pKa = 4.44FIHH272 pKa = 7.4DD273 pKa = 4.13GNPLGEE279 pKa = 4.28YY280 pKa = 10.96SNMEE284 pKa = 4.0YY285 pKa = 10.9EE286 pKa = 4.48LLNEE290 pKa = 4.48VSASTDD296 pKa = 3.38FSDD299 pKa = 3.5YY300 pKa = 11.01HH301 pKa = 7.75SVVDD305 pKa = 3.89EE306 pKa = 4.68DD307 pKa = 5.63LINFNDD313 pKa = 3.99DD314 pKa = 3.11VTLEE318 pKa = 4.2TQSMIAHH325 pKa = 6.92GGSLSEE331 pKa = 4.27IEE333 pKa = 4.17EE334 pKa = 4.39TGDD337 pKa = 3.66LSSDD341 pKa = 3.24VDD343 pKa = 3.74RR344 pKa = 11.84LLSSIEE350 pKa = 3.84TTPRR354 pKa = 11.84DD355 pKa = 3.48DD356 pKa = 3.26VVYY359 pKa = 10.56IRR361 pKa = 11.84GDD363 pKa = 3.33IILGEE368 pKa = 4.14DD369 pKa = 3.84EE370 pKa = 5.19IIIEE374 pKa = 4.18NGEE377 pKa = 4.12IILNNTDD384 pKa = 3.64EE385 pKa = 4.7LFEE388 pKa = 5.13DD389 pKa = 4.03VPEE392 pKa = 4.08MLEE395 pKa = 4.01NANLLEE401 pKa = 4.49DD402 pKa = 3.8RR403 pKa = 11.84EE404 pKa = 4.66HH405 pKa = 7.44VFEE408 pKa = 4.42YY409 pKa = 10.54NISVANEE416 pKa = 3.48NTTNQNSYY424 pKa = 10.71NFNAPQDD431 pKa = 4.02NTLTNYY437 pKa = 10.32NISRR441 pKa = 11.84FLQRR445 pKa = 11.84YY446 pKa = 6.69GTTTPNTQHH455 pKa = 6.75NISIHH460 pKa = 6.53RR461 pKa = 11.84DD462 pKa = 3.18GHH464 pKa = 5.2SLRR467 pKa = 11.84SDD469 pKa = 3.19TNIYY473 pKa = 10.86DD474 pKa = 4.61RR475 pKa = 11.84INGPSIRR482 pKa = 11.84EE483 pKa = 3.67NSLYY487 pKa = 10.28PGSFEE492 pKa = 3.77MAQYY496 pKa = 11.03ADD498 pKa = 4.72RR499 pKa = 11.84YY500 pKa = 10.15QMGHH504 pKa = 6.64TIAEE508 pKa = 4.43GFLYY512 pKa = 10.73
Molecular weight: 58.45 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A024VQE8|A0A024VQE8_PLAFA Uncharacterized protein OS=Plasmodium falciparum FCH/4 OX=1036724 GN=PFFCH_01616 PE=4 SV=1
MM1 pKa = 7.34VFPLTLLKK9 pKa = 10.59CSQNQPVMVEE19 pKa = 4.13LKK21 pKa = 10.8NGEE24 pKa = 4.4TYY26 pKa = 10.84SGFLVFCDD34 pKa = 2.98RR35 pKa = 11.84FMNLHH40 pKa = 6.2MKK42 pKa = 10.65NIICTSKK49 pKa = 11.12DD50 pKa = 2.75GDD52 pKa = 4.07KK53 pKa = 10.29FWKK56 pKa = 10.1ISEE59 pKa = 4.46CYY61 pKa = 10.25VRR63 pKa = 11.84GNSIKK68 pKa = 10.42YY69 pKa = 10.05IRR71 pKa = 11.84VQDD74 pKa = 3.68QAIEE78 pKa = 3.9QAIEE82 pKa = 3.64EE83 pKa = 4.28TAEE86 pKa = 3.88QKK88 pKa = 10.3ARR90 pKa = 11.84NMGRR94 pKa = 11.84GRR96 pKa = 11.84GGRR99 pKa = 11.84GRR101 pKa = 11.84GRR103 pKa = 11.84GGMNRR108 pKa = 11.84GTYY111 pKa = 9.58GRR113 pKa = 11.84GRR115 pKa = 11.84RR116 pKa = 11.84GSMRR120 pKa = 11.84RR121 pKa = 11.84GGRR124 pKa = 11.84GQQ126 pKa = 3.02
MM1 pKa = 7.34VFPLTLLKK9 pKa = 10.59CSQNQPVMVEE19 pKa = 4.13LKK21 pKa = 10.8NGEE24 pKa = 4.4TYY26 pKa = 10.84SGFLVFCDD34 pKa = 2.98RR35 pKa = 11.84FMNLHH40 pKa = 6.2MKK42 pKa = 10.65NIICTSKK49 pKa = 11.12DD50 pKa = 2.75GDD52 pKa = 4.07KK53 pKa = 10.29FWKK56 pKa = 10.1ISEE59 pKa = 4.46CYY61 pKa = 10.25VRR63 pKa = 11.84GNSIKK68 pKa = 10.42YY69 pKa = 10.05IRR71 pKa = 11.84VQDD74 pKa = 3.68QAIEE78 pKa = 3.9QAIEE82 pKa = 3.64EE83 pKa = 4.28TAEE86 pKa = 3.88QKK88 pKa = 10.3ARR90 pKa = 11.84NMGRR94 pKa = 11.84GRR96 pKa = 11.84GGRR99 pKa = 11.84GRR101 pKa = 11.84GRR103 pKa = 11.84GGMNRR108 pKa = 11.84GTYY111 pKa = 9.58GRR113 pKa = 11.84GRR115 pKa = 11.84RR116 pKa = 11.84GSMRR120 pKa = 11.84RR121 pKa = 11.84GGRR124 pKa = 11.84GQQ126 pKa = 3.02
Molecular weight: 14.3 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
3694402 |
9 |
10252 |
641.2 |
75.7 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
1.899 ± 0.022 | 1.79 ± 0.019 |
6.389 ± 0.027 | 6.961 ± 0.041 |
4.398 ± 0.025 | 2.766 ± 0.027 |
2.431 ± 0.015 | 9.357 ± 0.035 |
11.684 ± 0.042 | 7.597 ± 0.037 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.239 ± 0.014 | 14.731 ± 0.116 |
1.931 ± 0.02 | 2.739 ± 0.016 |
2.628 ± 0.019 | 6.396 ± 0.023 |
4.072 ± 0.019 | 3.727 ± 0.022 |
0.489 ± 0.008 | 5.776 ± 0.028 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
