
Rotavirus A (strain RVA/Cow/United Kingdom/UK/1975/G6P7[5]) (RV-A)
Taxonomy: Viruses; Riboviria; Orthornavirae; Duplornaviricota; Resentoviricetes; Reovirales; Reoviridae; Sedoreovirinae; Rotavirus; Rotavirus A; Bovine rotavirus A; Bovine rotavirus G6
Average proteome isoelectric point is 6.25
Get precalculated fractions of proteins
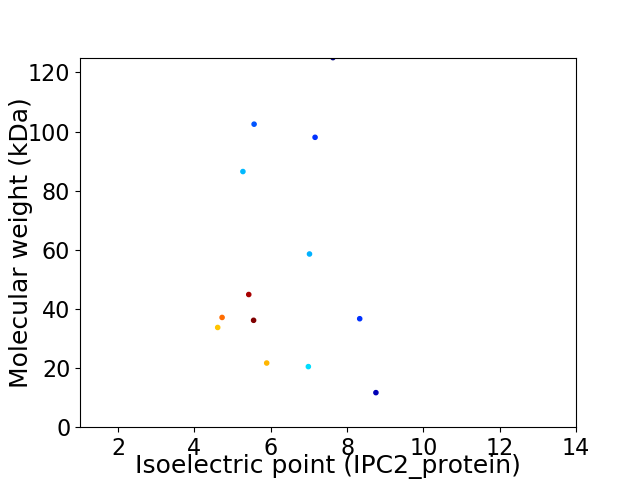
Virtual 2D-PAGE plot for 13 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
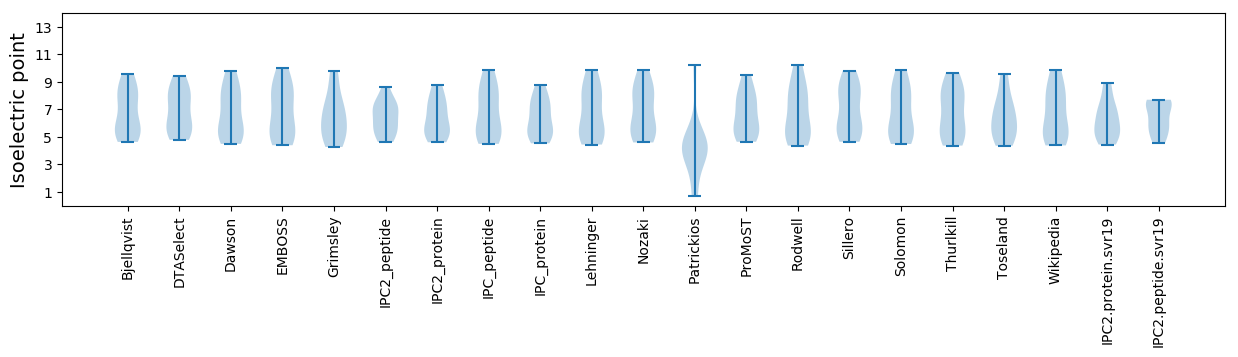
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P03534-2|VP7-2_ROTBU Isoform of P03534 Isoform 2 of Outer capsid glycoprotein VP7 OS=Rotavirus A (strain RVA/Cow/United Kingdom/UK/1975/G6P7[5]) OX=10934 PE=2 SV=1
MM1 pKa = 8.16DD2 pKa = 4.71YY3 pKa = 10.55IIYY6 pKa = 10.24RR7 pKa = 11.84FLLIVVVLATMINAQNYY24 pKa = 7.95GVNLPITGSMDD35 pKa = 3.06TAYY38 pKa = 10.66ANSTQSEE45 pKa = 4.42PFLTSTLCLYY55 pKa = 10.92YY56 pKa = 9.93PVEE59 pKa = 4.38ASNEE63 pKa = 3.97IADD66 pKa = 4.42TEE68 pKa = 4.41WKK70 pKa = 8.51DD71 pKa = 3.48TLSQLFLTKK80 pKa = 10.05GWPTGSVYY88 pKa = 10.44FKK90 pKa = 10.34EE91 pKa = 4.39YY92 pKa = 10.03TDD94 pKa = 3.19IAAFSVEE101 pKa = 3.86PQLYY105 pKa = 9.89CDD107 pKa = 3.87YY108 pKa = 11.21NLVLMKK114 pKa = 10.43YY115 pKa = 10.56DD116 pKa = 3.62STQEE120 pKa = 3.79LDD122 pKa = 3.4MSEE125 pKa = 4.16LADD128 pKa = 5.13LILNEE133 pKa = 4.24WLCNPMDD140 pKa = 3.29ITLYY144 pKa = 10.67YY145 pKa = 9.25YY146 pKa = 10.55QQTDD150 pKa = 3.57EE151 pKa = 4.17ANKK154 pKa = 8.08WISMGSSCTVKK165 pKa = 10.28VCPLNTQTLGIGCLITNPDD184 pKa = 3.37TFEE187 pKa = 4.14TVATTEE193 pKa = 4.21KK194 pKa = 10.92LVITDD199 pKa = 3.3VVDD202 pKa = 4.33GVNHH206 pKa = 6.76KK207 pKa = 11.09LNVTTATCTIRR218 pKa = 11.84NCKK221 pKa = 9.83KK222 pKa = 10.18LGPRR226 pKa = 11.84EE227 pKa = 3.94NVAIIQVGGANVLDD241 pKa = 3.77ITADD245 pKa = 3.5PTTAPQTEE253 pKa = 3.66RR254 pKa = 11.84MMRR257 pKa = 11.84INWKK261 pKa = 9.54KK262 pKa = 7.26WWQVFYY268 pKa = 10.66TVVDD272 pKa = 3.52YY273 pKa = 11.46VNQIIQTMSKK283 pKa = 10.22RR284 pKa = 11.84SRR286 pKa = 11.84SLNSSAFYY294 pKa = 11.0YY295 pKa = 10.28RR296 pKa = 11.84VV297 pKa = 3.04
MM1 pKa = 8.16DD2 pKa = 4.71YY3 pKa = 10.55IIYY6 pKa = 10.24RR7 pKa = 11.84FLLIVVVLATMINAQNYY24 pKa = 7.95GVNLPITGSMDD35 pKa = 3.06TAYY38 pKa = 10.66ANSTQSEE45 pKa = 4.42PFLTSTLCLYY55 pKa = 10.92YY56 pKa = 9.93PVEE59 pKa = 4.38ASNEE63 pKa = 3.97IADD66 pKa = 4.42TEE68 pKa = 4.41WKK70 pKa = 8.51DD71 pKa = 3.48TLSQLFLTKK80 pKa = 10.05GWPTGSVYY88 pKa = 10.44FKK90 pKa = 10.34EE91 pKa = 4.39YY92 pKa = 10.03TDD94 pKa = 3.19IAAFSVEE101 pKa = 3.86PQLYY105 pKa = 9.89CDD107 pKa = 3.87YY108 pKa = 11.21NLVLMKK114 pKa = 10.43YY115 pKa = 10.56DD116 pKa = 3.62STQEE120 pKa = 3.79LDD122 pKa = 3.4MSEE125 pKa = 4.16LADD128 pKa = 5.13LILNEE133 pKa = 4.24WLCNPMDD140 pKa = 3.29ITLYY144 pKa = 10.67YY145 pKa = 9.25YY146 pKa = 10.55QQTDD150 pKa = 3.57EE151 pKa = 4.17ANKK154 pKa = 8.08WISMGSSCTVKK165 pKa = 10.28VCPLNTQTLGIGCLITNPDD184 pKa = 3.37TFEE187 pKa = 4.14TVATTEE193 pKa = 4.21KK194 pKa = 10.92LVITDD199 pKa = 3.3VVDD202 pKa = 4.33GVNHH206 pKa = 6.76KK207 pKa = 11.09LNVTTATCTIRR218 pKa = 11.84NCKK221 pKa = 9.83KK222 pKa = 10.18LGPRR226 pKa = 11.84EE227 pKa = 3.94NVAIIQVGGANVLDD241 pKa = 3.77ITADD245 pKa = 3.5PTTAPQTEE253 pKa = 3.66RR254 pKa = 11.84MMRR257 pKa = 11.84INWKK261 pKa = 9.54KK262 pKa = 7.26WWQVFYY268 pKa = 10.66TVVDD272 pKa = 3.52YY273 pKa = 11.46VNQIIQTMSKK283 pKa = 10.22RR284 pKa = 11.84SRR286 pKa = 11.84SLNSSAFYY294 pKa = 11.0YY295 pKa = 10.28RR296 pKa = 11.84VV297 pKa = 3.04
Molecular weight: 33.73 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P12474|VP4_ROTBU Outer capsid protein VP4 OS=Rotavirus A (strain RVA/Cow/United Kingdom/UK/1975/G6P7[5]) OX=10934 PE=3 SV=1
MM1 pKa = 7.27NHH3 pKa = 6.21LQQRR7 pKa = 11.84QLFLEE12 pKa = 4.16NLLVGVNNMFHH23 pKa = 6.33QMQKK27 pKa = 10.4RR28 pKa = 11.84PVNTCCRR35 pKa = 11.84SLQKK39 pKa = 10.64ILDD42 pKa = 3.97HH43 pKa = 7.17LILLQTIHH51 pKa = 6.63SPAFRR56 pKa = 11.84LDD58 pKa = 3.53QMQLRR63 pKa = 11.84QMQTLACLWIHH74 pKa = 6.26QYY76 pKa = 10.72NHH78 pKa = 6.34DD79 pKa = 3.87HH80 pKa = 5.81QVTLGAIKK88 pKa = 9.95WISPLIKK95 pKa = 9.9EE96 pKa = 4.73LKK98 pKa = 10.03
MM1 pKa = 7.27NHH3 pKa = 6.21LQQRR7 pKa = 11.84QLFLEE12 pKa = 4.16NLLVGVNNMFHH23 pKa = 6.33QMQKK27 pKa = 10.4RR28 pKa = 11.84PVNTCCRR35 pKa = 11.84SLQKK39 pKa = 10.64ILDD42 pKa = 3.97HH43 pKa = 7.17LILLQTIHH51 pKa = 6.63SPAFRR56 pKa = 11.84LDD58 pKa = 3.53QMQLRR63 pKa = 11.84QMQTLACLWIHH74 pKa = 6.26QYY76 pKa = 10.72NHH78 pKa = 6.34DD79 pKa = 3.87HH80 pKa = 5.81QVTLGAIKK88 pKa = 9.95WISPLIKK95 pKa = 9.9EE96 pKa = 4.73LKK98 pKa = 10.03
Molecular weight: 11.69 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
6192 |
98 |
1088 |
476.3 |
54.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.346 ± 0.438 | 1.26 ± 0.329 |
5.975 ± 0.341 | 5.636 ± 0.288 |
4.167 ± 0.284 | 3.569 ± 0.356 |
1.486 ± 0.31 | 7.348 ± 0.378 |
6.411 ± 0.572 | 9.222 ± 0.472 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.198 ± 0.209 | 6.912 ± 0.321 |
3.23 ± 0.226 | 4.328 ± 0.482 |
4.797 ± 0.281 | 7.607 ± 0.827 |
6.395 ± 0.551 | 6.767 ± 0.297 |
1.211 ± 0.148 | 5.136 ± 0.471 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
