
Aquisalimonas asiatica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Chromatiales; Ectothiorhodospiraceae; Aquisalimonas
Average proteome isoelectric point is 5.91
Get precalculated fractions of proteins
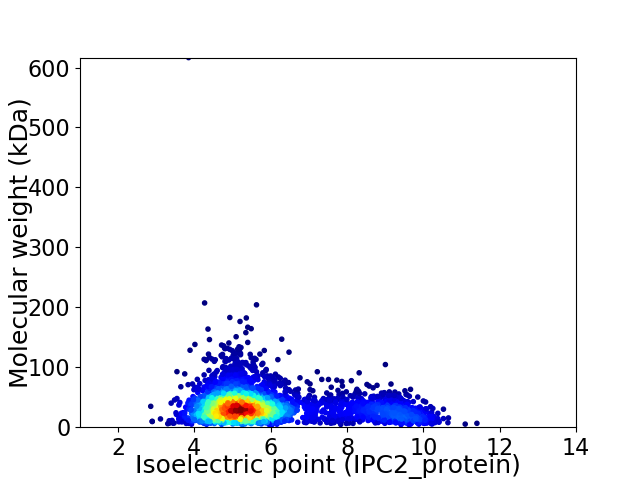
Virtual 2D-PAGE plot for 3698 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1H8SZY3|A0A1H8SZY3_9GAMM ADP-ribosylglycohydrolase OS=Aquisalimonas asiatica OX=406100 GN=SAMN04488052_103324 PE=4 SV=1
MM1 pKa = 6.49TTRR4 pKa = 11.84TSATTVFCIACASLLNGCFGGSSSGSGAGDD34 pKa = 4.1DD35 pKa = 3.89NDD37 pKa = 3.98GVQPEE42 pKa = 4.34DD43 pKa = 3.48VQVRR47 pKa = 11.84QFDD50 pKa = 3.82ASDD53 pKa = 3.37HH54 pKa = 6.85DD55 pKa = 4.4EE56 pKa = 4.37PVHH59 pKa = 7.08VDD61 pKa = 3.83LSTGEE66 pKa = 4.27VLTLSAGEE74 pKa = 4.28AADD77 pKa = 4.09SEE79 pKa = 4.47AWDD82 pKa = 3.38IALQRR87 pKa = 11.84HH88 pKa = 5.28QISLNGGTSGPGRR101 pKa = 11.84VQGALLDD108 pKa = 4.35GQASFYY114 pKa = 11.46NEE116 pKa = 4.84DD117 pKa = 3.46GTPNASVFTNTGAADD132 pKa = 3.64TLDD135 pKa = 4.08ALLAPVDD142 pKa = 4.23AVDD145 pKa = 4.01SGDD148 pKa = 3.42WVLDD152 pKa = 3.92SITTEE157 pKa = 5.38FGDD160 pKa = 3.26DD161 pKa = 2.86WYY163 pKa = 10.14IYY165 pKa = 11.03DD166 pKa = 4.01FATGEE171 pKa = 4.1FQPDD175 pKa = 3.9PDD177 pKa = 4.27NGWLLRR183 pKa = 11.84SGTGDD188 pKa = 2.56SYY190 pKa = 11.81ARR192 pKa = 11.84VRR194 pKa = 11.84ITEE197 pKa = 4.09LDD199 pKa = 3.37FDD201 pKa = 4.15SRR203 pKa = 11.84AGDD206 pKa = 3.6GVEE209 pKa = 3.83HH210 pKa = 6.59FRR212 pKa = 11.84FEE214 pKa = 4.75FDD216 pKa = 3.37VQVPNTQQFTEE227 pKa = 4.36TAVFTGSLPGGGGDD241 pKa = 3.68LCFDD245 pKa = 3.84VDD247 pKa = 4.03SDD249 pKa = 3.87ATADD253 pKa = 3.72CSGVDD258 pKa = 3.22WDD260 pKa = 4.26IQIGFQGQDD269 pKa = 2.75FYY271 pKa = 11.83LRR273 pKa = 11.84SNSGVSGDD281 pKa = 3.36GDD283 pKa = 3.47GGVFGPFPWSEE294 pKa = 3.9LEE296 pKa = 4.58AYY298 pKa = 10.23DD299 pKa = 4.2SATVTPSGEE308 pKa = 4.43SIAGHH313 pKa = 5.34YY314 pKa = 10.01QPDD317 pKa = 3.76ASTGIFDD324 pKa = 4.23AYY326 pKa = 10.58SWYY329 pKa = 10.54AYY331 pKa = 10.87NLTGQHH337 pKa = 5.6QLWPNYY343 pKa = 8.99RR344 pKa = 11.84VYY346 pKa = 11.03LIDD349 pKa = 4.35SDD351 pKa = 4.24HH352 pKa = 7.44DD353 pKa = 3.73EE354 pKa = 4.7DD355 pKa = 5.52AAAQYY360 pKa = 11.08ALQVINYY367 pKa = 8.91YY368 pKa = 10.83DD369 pKa = 3.48GTGDD373 pKa = 3.37SGHH376 pKa = 7.27PEE378 pKa = 4.0LRR380 pKa = 11.84WRR382 pKa = 11.84QVDD385 pKa = 3.22
MM1 pKa = 6.49TTRR4 pKa = 11.84TSATTVFCIACASLLNGCFGGSSSGSGAGDD34 pKa = 4.1DD35 pKa = 3.89NDD37 pKa = 3.98GVQPEE42 pKa = 4.34DD43 pKa = 3.48VQVRR47 pKa = 11.84QFDD50 pKa = 3.82ASDD53 pKa = 3.37HH54 pKa = 6.85DD55 pKa = 4.4EE56 pKa = 4.37PVHH59 pKa = 7.08VDD61 pKa = 3.83LSTGEE66 pKa = 4.27VLTLSAGEE74 pKa = 4.28AADD77 pKa = 4.09SEE79 pKa = 4.47AWDD82 pKa = 3.38IALQRR87 pKa = 11.84HH88 pKa = 5.28QISLNGGTSGPGRR101 pKa = 11.84VQGALLDD108 pKa = 4.35GQASFYY114 pKa = 11.46NEE116 pKa = 4.84DD117 pKa = 3.46GTPNASVFTNTGAADD132 pKa = 3.64TLDD135 pKa = 4.08ALLAPVDD142 pKa = 4.23AVDD145 pKa = 4.01SGDD148 pKa = 3.42WVLDD152 pKa = 3.92SITTEE157 pKa = 5.38FGDD160 pKa = 3.26DD161 pKa = 2.86WYY163 pKa = 10.14IYY165 pKa = 11.03DD166 pKa = 4.01FATGEE171 pKa = 4.1FQPDD175 pKa = 3.9PDD177 pKa = 4.27NGWLLRR183 pKa = 11.84SGTGDD188 pKa = 2.56SYY190 pKa = 11.81ARR192 pKa = 11.84VRR194 pKa = 11.84ITEE197 pKa = 4.09LDD199 pKa = 3.37FDD201 pKa = 4.15SRR203 pKa = 11.84AGDD206 pKa = 3.6GVEE209 pKa = 3.83HH210 pKa = 6.59FRR212 pKa = 11.84FEE214 pKa = 4.75FDD216 pKa = 3.37VQVPNTQQFTEE227 pKa = 4.36TAVFTGSLPGGGGDD241 pKa = 3.68LCFDD245 pKa = 3.84VDD247 pKa = 4.03SDD249 pKa = 3.87ATADD253 pKa = 3.72CSGVDD258 pKa = 3.22WDD260 pKa = 4.26IQIGFQGQDD269 pKa = 2.75FYY271 pKa = 11.83LRR273 pKa = 11.84SNSGVSGDD281 pKa = 3.36GDD283 pKa = 3.47GGVFGPFPWSEE294 pKa = 3.9LEE296 pKa = 4.58AYY298 pKa = 10.23DD299 pKa = 4.2SATVTPSGEE308 pKa = 4.43SIAGHH313 pKa = 5.34YY314 pKa = 10.01QPDD317 pKa = 3.76ASTGIFDD324 pKa = 4.23AYY326 pKa = 10.58SWYY329 pKa = 10.54AYY331 pKa = 10.87NLTGQHH337 pKa = 5.6QLWPNYY343 pKa = 8.99RR344 pKa = 11.84VYY346 pKa = 11.03LIDD349 pKa = 4.35SDD351 pKa = 4.24HH352 pKa = 7.44DD353 pKa = 3.73EE354 pKa = 4.7DD355 pKa = 5.52AAAQYY360 pKa = 11.08ALQVINYY367 pKa = 8.91YY368 pKa = 10.83DD369 pKa = 3.48GTGDD373 pKa = 3.37SGHH376 pKa = 7.27PEE378 pKa = 4.0LRR380 pKa = 11.84WRR382 pKa = 11.84QVDD385 pKa = 3.22
Molecular weight: 41.28 kDa
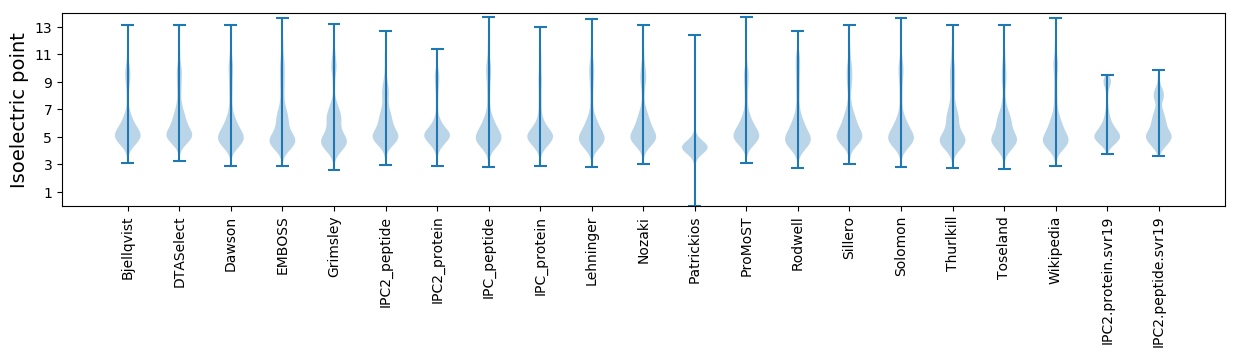
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1H8VD67|A0A1H8VD67_9GAMM Uncharacterized protein OS=Aquisalimonas asiatica OX=406100 GN=SAMN04488052_11139 PE=4 SV=1
MM1 pKa = 7.12FRR3 pKa = 11.84MFRR6 pKa = 11.84MFRR9 pKa = 11.84MFRR12 pKa = 11.84MFRR15 pKa = 11.84MFRR18 pKa = 11.84MFRR21 pKa = 11.84MFRR24 pKa = 11.84MFRR27 pKa = 11.84MFRR30 pKa = 11.84VVRR33 pKa = 11.84VVRR36 pKa = 11.84VVRR39 pKa = 11.84VVRR42 pKa = 11.84VVRR45 pKa = 11.84VVRR48 pKa = 3.93
MM1 pKa = 7.12FRR3 pKa = 11.84MFRR6 pKa = 11.84MFRR9 pKa = 11.84MFRR12 pKa = 11.84MFRR15 pKa = 11.84MFRR18 pKa = 11.84MFRR21 pKa = 11.84MFRR24 pKa = 11.84MFRR27 pKa = 11.84MFRR30 pKa = 11.84VVRR33 pKa = 11.84VVRR36 pKa = 11.84VVRR39 pKa = 11.84VVRR42 pKa = 11.84VVRR45 pKa = 11.84VVRR48 pKa = 3.93
Molecular weight: 6.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1223554 |
40 |
6102 |
330.9 |
36.22 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.413 ± 0.056 | 0.854 ± 0.012 |
6.564 ± 0.045 | 6.473 ± 0.039 |
3.373 ± 0.028 | 8.741 ± 0.041 |
2.411 ± 0.021 | 4.332 ± 0.031 |
1.977 ± 0.03 | 10.519 ± 0.053 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.491 ± 0.02 | 2.469 ± 0.025 |
5.036 ± 0.032 | 3.616 ± 0.025 |
7.969 ± 0.043 | 4.961 ± 0.028 |
5.184 ± 0.027 | 7.783 ± 0.035 |
1.453 ± 0.018 | 2.38 ± 0.019 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
