
Lacinutrix sp. CAU 1491
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Lacinutrix; unclassified Lacinutrix
Average proteome isoelectric point is 6.76
Get precalculated fractions of proteins
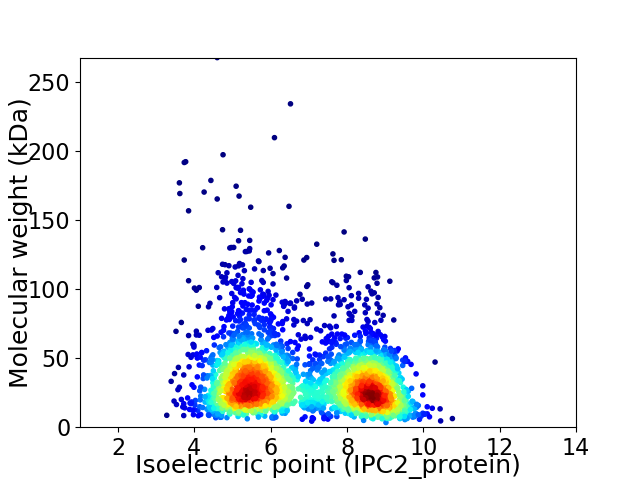
Virtual 2D-PAGE plot for 2986 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4U0EVL1|A0A4U0EVL1_9FLAO TetR/AcrR family transcriptional regulator OS=Lacinutrix sp. CAU 1491 OX=2565367 GN=E5167_08800 PE=4 SV=1
MM1 pKa = 7.65KK2 pKa = 9.11KK3 pKa = 6.63TTYY6 pKa = 10.84SLYY9 pKa = 10.37HH10 pKa = 5.94WMCILLLLVSGSLYY24 pKa = 10.08SQTVIDD30 pKa = 4.74SEE32 pKa = 4.55DD33 pKa = 3.85FEE35 pKa = 5.19SGWGIWNDD43 pKa = 3.45GGSDD47 pKa = 3.64CFLNTGSILSGANSVNLQDD66 pKa = 5.43NNNQDD71 pKa = 2.79SSMYY75 pKa = 10.58TNDD78 pKa = 3.31IDD80 pKa = 3.9LTTFGSVTIVFDD92 pKa = 4.12FRR94 pKa = 11.84TSGFNNGHH102 pKa = 6.72DD103 pKa = 3.8FFIEE107 pKa = 4.44FSNNGGASWNATPITTYY124 pKa = 10.68VRR126 pKa = 11.84GSNFNNDD133 pKa = 2.78TTYY136 pKa = 10.5TGEE139 pKa = 4.15TATITAGGTYY149 pKa = 9.72TFTANSRR156 pKa = 11.84FRR158 pKa = 11.84FRR160 pKa = 11.84ADD162 pKa = 3.0ASQSSDD168 pKa = 3.57DD169 pKa = 4.54LYY171 pKa = 10.86IDD173 pKa = 3.56NVVMTGYY180 pKa = 10.06PKK182 pKa = 10.25TPEE185 pKa = 3.69INITGLGNTINDD197 pKa = 3.67GDD199 pKa = 4.19TTPSVTDD206 pKa = 3.38DD207 pKa = 3.51TDD209 pKa = 3.72FGTVNVAGGTNPNTFTIQNTGTAALSLTGASPYY242 pKa = 9.37VTISGTNAADD252 pKa = 3.86FAVTTAPANTIASSGSTTFVITFDD276 pKa = 3.76PSAAGIRR283 pKa = 11.84TATVSIANDD292 pKa = 3.55DD293 pKa = 4.29SNEE296 pKa = 3.75NPYY299 pKa = 11.21NFDD302 pKa = 3.29IEE304 pKa = 4.72GNGYY308 pKa = 10.18VPVPEE313 pKa = 4.99INVQGNSVDD322 pKa = 4.56IVDD325 pKa = 4.64GDD327 pKa = 3.92NTPSLTDD334 pKa = 3.16ATDD337 pKa = 3.32YY338 pKa = 9.61GTVNVEE344 pKa = 4.18SLVSRR349 pKa = 11.84TFTIQNTGTADD360 pKa = 3.56LTISSITLSNTTDD373 pKa = 3.71FSIIGTPYY381 pKa = 9.81ASPVASSGSTTFTVQFNSLTIGTKK405 pKa = 6.92TTTVTINNNDD415 pKa = 3.64SDD417 pKa = 3.83EE418 pKa = 4.25STYY421 pKa = 11.03QFDD424 pKa = 3.4IEE426 pKa = 5.3ARR428 pKa = 11.84AEE430 pKa = 4.09QNFFDD435 pKa = 4.39SDD437 pKa = 3.39GDD439 pKa = 4.18GIYY442 pKa = 10.96DD443 pKa = 5.19NIDD446 pKa = 3.01IDD448 pKa = 4.97DD449 pKa = 5.02DD450 pKa = 4.29NDD452 pKa = 4.8GILDD456 pKa = 3.84SEE458 pKa = 4.85EE459 pKa = 4.09EE460 pKa = 4.15LACQVSSTSTVMNYY474 pKa = 10.27KK475 pKa = 10.13FLNEE479 pKa = 4.12TFGEE483 pKa = 4.68GTNRR487 pKa = 11.84TTINTTYY494 pKa = 11.03DD495 pKa = 2.87ATTTYY500 pKa = 10.35CYY502 pKa = 10.71EE503 pKa = 5.29DD504 pKa = 3.81GTASCPSLGGIDD516 pKa = 4.94LNDD519 pKa = 3.47GEE521 pKa = 4.57YY522 pKa = 10.82AVYY525 pKa = 8.74YY526 pKa = 9.0TAGTNDD532 pKa = 3.46GTNNTPNGEE541 pKa = 4.08VASWVDD547 pKa = 3.39GLWYY551 pKa = 9.57TGEE554 pKa = 4.21DD555 pKa = 3.46HH556 pKa = 7.58TSGDD560 pKa = 3.78TNGRR564 pKa = 11.84MAMFNASYY572 pKa = 11.07DD573 pKa = 3.3PGTFYY578 pKa = 10.62TATITGALPNVPVTYY593 pKa = 10.62SFWVLNLDD601 pKa = 3.46ISTANNINTRR611 pKa = 11.84IRR613 pKa = 11.84PNILVEE619 pKa = 4.16FRR621 pKa = 11.84DD622 pKa = 3.4AGGNVLQSITTGDD635 pKa = 3.56IPPSIEE641 pKa = 3.93GDD643 pKa = 3.63PANSWHH649 pKa = 6.46QFTADD654 pKa = 3.09LTLNVSEE661 pKa = 4.97FYY663 pKa = 11.17VYY665 pKa = 10.18FINNNPGGLGNDD677 pKa = 3.81LAIDD681 pKa = 5.14DD682 pKa = 4.33IVISQTLCDD691 pKa = 3.97TDD693 pKa = 3.4NDD695 pKa = 4.78GIADD699 pKa = 3.99VFDD702 pKa = 5.4LDD704 pKa = 4.48SDD706 pKa = 3.9NDD708 pKa = 4.68GIPDD712 pKa = 3.57VVEE715 pKa = 4.74AGLGDD720 pKa = 4.12LSEE723 pKa = 4.62GTATLTGIASWVDD736 pKa = 3.19ANGNGMHH743 pKa = 7.34DD744 pKa = 3.71LAEE747 pKa = 4.71GNSVLDD753 pKa = 3.58TDD755 pKa = 4.52GDD757 pKa = 4.29GVPNYY762 pKa = 10.71LDD764 pKa = 4.89LDD766 pKa = 4.08SDD768 pKa = 3.73NDD770 pKa = 4.28TIFDD774 pKa = 3.71VDD776 pKa = 3.38EE777 pKa = 4.83SGAGNTADD785 pKa = 4.17PSYY788 pKa = 11.4QNGDD792 pKa = 2.87GDD794 pKa = 3.75ITGDD798 pKa = 3.82GVGDD802 pKa = 4.29GPDD805 pKa = 3.11TDD807 pKa = 5.56AIRR810 pKa = 11.84EE811 pKa = 3.98TDD813 pKa = 3.19IDD815 pKa = 3.92SDD817 pKa = 3.88GTSEE821 pKa = 4.23YY822 pKa = 9.81FTDD825 pKa = 6.12GILDD829 pKa = 3.35IYY831 pKa = 10.96DD832 pKa = 3.85YY833 pKa = 11.72YY834 pKa = 11.75NGGTFGTAYY843 pKa = 10.48GNSNQGLGNTYY854 pKa = 10.54YY855 pKa = 11.31VLDD858 pKa = 4.47SDD860 pKa = 4.72SDD862 pKa = 4.35GIPDD866 pKa = 3.7YY867 pKa = 11.04MDD869 pKa = 3.17ITSDD873 pKa = 3.25GVTFDD878 pKa = 4.11ISHH881 pKa = 6.79TLYY884 pKa = 11.37ANLDD888 pKa = 3.77ANNDD892 pKa = 3.56GVIDD896 pKa = 3.99DD897 pKa = 4.71TNDD900 pKa = 3.36SEE902 pKa = 5.42GDD904 pKa = 3.87GILDD908 pKa = 5.35LFDD911 pKa = 4.13TDD913 pKa = 4.42DD914 pKa = 3.99AAFGSPRR921 pKa = 11.84DD922 pKa = 3.77LDD924 pKa = 4.11RR925 pKa = 11.84KK926 pKa = 9.12LHH928 pKa = 6.43LYY930 pKa = 10.35FDD932 pKa = 3.92GRR934 pKa = 11.84NDD936 pKa = 3.92YY937 pKa = 11.02IEE939 pKa = 4.68DD940 pKa = 3.61ASVIGTWSEE949 pKa = 3.72ATQMGWIKK957 pKa = 10.21IDD959 pKa = 3.38PTITGGSKK967 pKa = 9.68TIMGQDD973 pKa = 2.96NFYY976 pKa = 9.53ITLNADD982 pKa = 3.91LSISATANGTTITNGGALATDD1003 pKa = 3.81RR1004 pKa = 11.84WVHH1007 pKa = 6.55IGATYY1012 pKa = 11.2SNIDD1016 pKa = 4.13NIFKK1020 pKa = 10.78LYY1022 pKa = 10.48INGLEE1027 pKa = 4.19VSSSAISGSLPSDD1040 pKa = 3.29TSSYY1044 pKa = 10.46TLGRR1048 pKa = 11.84NPDD1051 pKa = 3.33TDD1053 pKa = 2.84SGYY1056 pKa = 10.72FMGYY1060 pKa = 8.68IDD1062 pKa = 3.85EE1063 pKa = 5.28AKK1065 pKa = 10.81VFDD1068 pKa = 4.17KK1069 pKa = 11.22ALTADD1074 pKa = 4.37EE1075 pKa = 4.3YY1076 pKa = 11.32QKK1078 pKa = 11.19NVYY1081 pKa = 9.26QEE1083 pKa = 4.68IEE1085 pKa = 4.44DD1086 pKa = 3.72NTLVRR1091 pKa = 11.84GAIIPKK1097 pKa = 10.31NITTLSWSNVQRR1109 pKa = 11.84YY1110 pKa = 7.47FRR1112 pKa = 11.84FDD1114 pKa = 3.18VYY1116 pKa = 11.2KK1117 pKa = 10.96DD1118 pKa = 4.66DD1119 pKa = 3.93ITDD1122 pKa = 4.09DD1123 pKa = 3.98LTTGTIDD1130 pKa = 3.05TGTGAKK1136 pKa = 8.98MYY1138 pKa = 9.81NFKK1141 pKa = 10.75IIDD1144 pKa = 3.85YY1145 pKa = 10.22QSAPLPFVTQQSGSLPTAVDD1165 pKa = 3.23IASDD1169 pKa = 4.03GVNGADD1175 pKa = 4.43AITYY1179 pKa = 7.95DD1180 pKa = 3.25WSIVRR1185 pKa = 11.84VEE1187 pKa = 4.82HH1188 pKa = 7.94DD1189 pKa = 4.66DD1190 pKa = 2.88ITYY1193 pKa = 10.77NDD1195 pKa = 3.92RR1196 pKa = 11.84QKK1198 pKa = 11.29HH1199 pKa = 5.45LGLFVNEE1206 pKa = 4.69LDD1208 pKa = 4.37AGLNPIEE1215 pKa = 5.24FSVQNDD1221 pKa = 3.58SEE1223 pKa = 5.83LNVSWYY1229 pKa = 10.86LKK1231 pKa = 10.71LDD1233 pKa = 3.44GLIDD1237 pKa = 4.62LEE1239 pKa = 4.89GEE1241 pKa = 4.26SQLVQGSDD1249 pKa = 3.08STLDD1253 pKa = 3.37INSKK1257 pKa = 8.7GTIEE1261 pKa = 4.9RR1262 pKa = 11.84DD1263 pKa = 3.49QQGTADD1269 pKa = 4.48LYY1271 pKa = 10.78TYY1273 pKa = 9.98NYY1275 pKa = 7.23WAAPVGVTSTISNNNNYY1292 pKa = 7.81TLPDD1296 pKa = 3.65VLNDD1300 pKa = 3.52GTTPAIPSSINWLTSGYY1317 pKa = 10.23DD1318 pKa = 3.55GTSGSPVGIADD1329 pKa = 3.37YY1330 pKa = 9.81WIWKK1334 pKa = 7.23YY1335 pKa = 11.44ANRR1338 pKa = 11.84VSDD1341 pKa = 5.27NYY1343 pKa = 10.66PSWQHH1348 pKa = 4.87VRR1350 pKa = 11.84STGSLQPGEE1359 pKa = 4.69GYY1361 pKa = 8.82TMKK1364 pKa = 10.21GTTNTSGAIFTEE1376 pKa = 3.92QNYY1379 pKa = 8.94VFNGKK1384 pKa = 7.73PHH1386 pKa = 6.59NGDD1389 pKa = 2.72ITLTLSAGNDD1399 pKa = 3.55YY1400 pKa = 11.19LIGNPYY1406 pKa = 10.72ASAIDD1411 pKa = 3.71ANEE1414 pKa = 4.58FILDD1418 pKa = 4.36NISDD1422 pKa = 4.01GAGRR1426 pKa = 11.84AGSNIINGTLYY1437 pKa = 10.28FWEE1440 pKa = 4.83HH1441 pKa = 6.05FASSTHH1447 pKa = 6.26ILSEE1451 pKa = 4.08YY1452 pKa = 8.82QGGYY1456 pKa = 8.85GAYY1459 pKa = 9.19TLITGTAAINNDD1471 pKa = 2.61VRR1473 pKa = 11.84INHH1476 pKa = 6.37TGGLTSSKK1484 pKa = 10.2GAPEE1488 pKa = 4.02RR1489 pKa = 11.84YY1490 pKa = 9.41IPVAQGFFVTADD1502 pKa = 3.18TGGSITFKK1510 pKa = 10.55NSQRR1514 pKa = 11.84TFKK1517 pKa = 10.67TEE1519 pKa = 3.56AADD1522 pKa = 3.42PSVFMRR1528 pKa = 11.84VNDD1531 pKa = 4.21GKK1533 pKa = 11.19SKK1535 pKa = 9.96QKK1537 pKa = 10.3KK1538 pKa = 9.73SSVTTSKK1545 pKa = 7.72TTNDD1549 pKa = 2.84TRR1551 pKa = 11.84EE1552 pKa = 4.07VIKK1555 pKa = 10.78LYY1557 pKa = 10.53FDD1559 pKa = 3.63SPKK1562 pKa = 10.54GYY1564 pKa = 9.74HH1565 pKa = 5.53RR1566 pKa = 11.84QIAVGIDD1573 pKa = 3.17SNTTDD1578 pKa = 3.4GHH1580 pKa = 6.88DD1581 pKa = 3.27IGYY1584 pKa = 7.99DD1585 pKa = 3.21APLIEE1590 pKa = 5.95DD1591 pKa = 3.63NVEE1594 pKa = 3.9DD1595 pKa = 4.48MFWTFDD1601 pKa = 3.36NNKK1604 pKa = 9.42FVIQGVSDD1612 pKa = 3.71FNINRR1617 pKa = 11.84TLPLGVKK1624 pKa = 9.32IDD1626 pKa = 3.81QEE1628 pKa = 4.39GTATIRR1634 pKa = 11.84VDD1636 pKa = 3.52YY1637 pKa = 10.8LEE1639 pKa = 5.54NIPIDD1644 pKa = 3.75TNIYY1648 pKa = 10.54LLDD1651 pKa = 4.35KK1652 pKa = 10.47EE1653 pKa = 4.5LEE1655 pKa = 4.17IYY1657 pKa = 10.53HH1658 pKa = 6.96DD1659 pKa = 3.99LRR1661 pKa = 11.84EE1662 pKa = 4.29SNYY1665 pKa = 10.41EE1666 pKa = 3.78VFLTIGEE1673 pKa = 4.27HH1674 pKa = 5.44YY1675 pKa = 10.79DD1676 pKa = 3.14RR1677 pKa = 11.84FEE1679 pKa = 3.87ITFSNGEE1686 pKa = 4.14EE1687 pKa = 3.93EE1688 pKa = 4.11VDD1690 pKa = 4.39PIEE1693 pKa = 5.42GLTKK1697 pKa = 10.54TNLHH1701 pKa = 6.57FSNDD1705 pKa = 3.35KK1706 pKa = 10.77EE1707 pKa = 4.56SIIVINPDD1715 pKa = 2.98NLIINSVKK1723 pKa = 9.59MVNILGQSVFKK1734 pKa = 10.35MDD1736 pKa = 3.2TAFEE1740 pKa = 4.12DD1741 pKa = 4.21RR1742 pKa = 11.84YY1743 pKa = 11.16KK1744 pKa = 10.88EE1745 pKa = 3.76ITVKK1749 pKa = 10.55HH1750 pKa = 6.4LNTGAYY1756 pKa = 8.64IINMITNKK1764 pKa = 10.44GLITKK1769 pKa = 9.9KK1770 pKa = 10.7VVVEE1774 pKa = 4.07
MM1 pKa = 7.65KK2 pKa = 9.11KK3 pKa = 6.63TTYY6 pKa = 10.84SLYY9 pKa = 10.37HH10 pKa = 5.94WMCILLLLVSGSLYY24 pKa = 10.08SQTVIDD30 pKa = 4.74SEE32 pKa = 4.55DD33 pKa = 3.85FEE35 pKa = 5.19SGWGIWNDD43 pKa = 3.45GGSDD47 pKa = 3.64CFLNTGSILSGANSVNLQDD66 pKa = 5.43NNNQDD71 pKa = 2.79SSMYY75 pKa = 10.58TNDD78 pKa = 3.31IDD80 pKa = 3.9LTTFGSVTIVFDD92 pKa = 4.12FRR94 pKa = 11.84TSGFNNGHH102 pKa = 6.72DD103 pKa = 3.8FFIEE107 pKa = 4.44FSNNGGASWNATPITTYY124 pKa = 10.68VRR126 pKa = 11.84GSNFNNDD133 pKa = 2.78TTYY136 pKa = 10.5TGEE139 pKa = 4.15TATITAGGTYY149 pKa = 9.72TFTANSRR156 pKa = 11.84FRR158 pKa = 11.84FRR160 pKa = 11.84ADD162 pKa = 3.0ASQSSDD168 pKa = 3.57DD169 pKa = 4.54LYY171 pKa = 10.86IDD173 pKa = 3.56NVVMTGYY180 pKa = 10.06PKK182 pKa = 10.25TPEE185 pKa = 3.69INITGLGNTINDD197 pKa = 3.67GDD199 pKa = 4.19TTPSVTDD206 pKa = 3.38DD207 pKa = 3.51TDD209 pKa = 3.72FGTVNVAGGTNPNTFTIQNTGTAALSLTGASPYY242 pKa = 9.37VTISGTNAADD252 pKa = 3.86FAVTTAPANTIASSGSTTFVITFDD276 pKa = 3.76PSAAGIRR283 pKa = 11.84TATVSIANDD292 pKa = 3.55DD293 pKa = 4.29SNEE296 pKa = 3.75NPYY299 pKa = 11.21NFDD302 pKa = 3.29IEE304 pKa = 4.72GNGYY308 pKa = 10.18VPVPEE313 pKa = 4.99INVQGNSVDD322 pKa = 4.56IVDD325 pKa = 4.64GDD327 pKa = 3.92NTPSLTDD334 pKa = 3.16ATDD337 pKa = 3.32YY338 pKa = 9.61GTVNVEE344 pKa = 4.18SLVSRR349 pKa = 11.84TFTIQNTGTADD360 pKa = 3.56LTISSITLSNTTDD373 pKa = 3.71FSIIGTPYY381 pKa = 9.81ASPVASSGSTTFTVQFNSLTIGTKK405 pKa = 6.92TTTVTINNNDD415 pKa = 3.64SDD417 pKa = 3.83EE418 pKa = 4.25STYY421 pKa = 11.03QFDD424 pKa = 3.4IEE426 pKa = 5.3ARR428 pKa = 11.84AEE430 pKa = 4.09QNFFDD435 pKa = 4.39SDD437 pKa = 3.39GDD439 pKa = 4.18GIYY442 pKa = 10.96DD443 pKa = 5.19NIDD446 pKa = 3.01IDD448 pKa = 4.97DD449 pKa = 5.02DD450 pKa = 4.29NDD452 pKa = 4.8GILDD456 pKa = 3.84SEE458 pKa = 4.85EE459 pKa = 4.09EE460 pKa = 4.15LACQVSSTSTVMNYY474 pKa = 10.27KK475 pKa = 10.13FLNEE479 pKa = 4.12TFGEE483 pKa = 4.68GTNRR487 pKa = 11.84TTINTTYY494 pKa = 11.03DD495 pKa = 2.87ATTTYY500 pKa = 10.35CYY502 pKa = 10.71EE503 pKa = 5.29DD504 pKa = 3.81GTASCPSLGGIDD516 pKa = 4.94LNDD519 pKa = 3.47GEE521 pKa = 4.57YY522 pKa = 10.82AVYY525 pKa = 8.74YY526 pKa = 9.0TAGTNDD532 pKa = 3.46GTNNTPNGEE541 pKa = 4.08VASWVDD547 pKa = 3.39GLWYY551 pKa = 9.57TGEE554 pKa = 4.21DD555 pKa = 3.46HH556 pKa = 7.58TSGDD560 pKa = 3.78TNGRR564 pKa = 11.84MAMFNASYY572 pKa = 11.07DD573 pKa = 3.3PGTFYY578 pKa = 10.62TATITGALPNVPVTYY593 pKa = 10.62SFWVLNLDD601 pKa = 3.46ISTANNINTRR611 pKa = 11.84IRR613 pKa = 11.84PNILVEE619 pKa = 4.16FRR621 pKa = 11.84DD622 pKa = 3.4AGGNVLQSITTGDD635 pKa = 3.56IPPSIEE641 pKa = 3.93GDD643 pKa = 3.63PANSWHH649 pKa = 6.46QFTADD654 pKa = 3.09LTLNVSEE661 pKa = 4.97FYY663 pKa = 11.17VYY665 pKa = 10.18FINNNPGGLGNDD677 pKa = 3.81LAIDD681 pKa = 5.14DD682 pKa = 4.33IVISQTLCDD691 pKa = 3.97TDD693 pKa = 3.4NDD695 pKa = 4.78GIADD699 pKa = 3.99VFDD702 pKa = 5.4LDD704 pKa = 4.48SDD706 pKa = 3.9NDD708 pKa = 4.68GIPDD712 pKa = 3.57VVEE715 pKa = 4.74AGLGDD720 pKa = 4.12LSEE723 pKa = 4.62GTATLTGIASWVDD736 pKa = 3.19ANGNGMHH743 pKa = 7.34DD744 pKa = 3.71LAEE747 pKa = 4.71GNSVLDD753 pKa = 3.58TDD755 pKa = 4.52GDD757 pKa = 4.29GVPNYY762 pKa = 10.71LDD764 pKa = 4.89LDD766 pKa = 4.08SDD768 pKa = 3.73NDD770 pKa = 4.28TIFDD774 pKa = 3.71VDD776 pKa = 3.38EE777 pKa = 4.83SGAGNTADD785 pKa = 4.17PSYY788 pKa = 11.4QNGDD792 pKa = 2.87GDD794 pKa = 3.75ITGDD798 pKa = 3.82GVGDD802 pKa = 4.29GPDD805 pKa = 3.11TDD807 pKa = 5.56AIRR810 pKa = 11.84EE811 pKa = 3.98TDD813 pKa = 3.19IDD815 pKa = 3.92SDD817 pKa = 3.88GTSEE821 pKa = 4.23YY822 pKa = 9.81FTDD825 pKa = 6.12GILDD829 pKa = 3.35IYY831 pKa = 10.96DD832 pKa = 3.85YY833 pKa = 11.72YY834 pKa = 11.75NGGTFGTAYY843 pKa = 10.48GNSNQGLGNTYY854 pKa = 10.54YY855 pKa = 11.31VLDD858 pKa = 4.47SDD860 pKa = 4.72SDD862 pKa = 4.35GIPDD866 pKa = 3.7YY867 pKa = 11.04MDD869 pKa = 3.17ITSDD873 pKa = 3.25GVTFDD878 pKa = 4.11ISHH881 pKa = 6.79TLYY884 pKa = 11.37ANLDD888 pKa = 3.77ANNDD892 pKa = 3.56GVIDD896 pKa = 3.99DD897 pKa = 4.71TNDD900 pKa = 3.36SEE902 pKa = 5.42GDD904 pKa = 3.87GILDD908 pKa = 5.35LFDD911 pKa = 4.13TDD913 pKa = 4.42DD914 pKa = 3.99AAFGSPRR921 pKa = 11.84DD922 pKa = 3.77LDD924 pKa = 4.11RR925 pKa = 11.84KK926 pKa = 9.12LHH928 pKa = 6.43LYY930 pKa = 10.35FDD932 pKa = 3.92GRR934 pKa = 11.84NDD936 pKa = 3.92YY937 pKa = 11.02IEE939 pKa = 4.68DD940 pKa = 3.61ASVIGTWSEE949 pKa = 3.72ATQMGWIKK957 pKa = 10.21IDD959 pKa = 3.38PTITGGSKK967 pKa = 9.68TIMGQDD973 pKa = 2.96NFYY976 pKa = 9.53ITLNADD982 pKa = 3.91LSISATANGTTITNGGALATDD1003 pKa = 3.81RR1004 pKa = 11.84WVHH1007 pKa = 6.55IGATYY1012 pKa = 11.2SNIDD1016 pKa = 4.13NIFKK1020 pKa = 10.78LYY1022 pKa = 10.48INGLEE1027 pKa = 4.19VSSSAISGSLPSDD1040 pKa = 3.29TSSYY1044 pKa = 10.46TLGRR1048 pKa = 11.84NPDD1051 pKa = 3.33TDD1053 pKa = 2.84SGYY1056 pKa = 10.72FMGYY1060 pKa = 8.68IDD1062 pKa = 3.85EE1063 pKa = 5.28AKK1065 pKa = 10.81VFDD1068 pKa = 4.17KK1069 pKa = 11.22ALTADD1074 pKa = 4.37EE1075 pKa = 4.3YY1076 pKa = 11.32QKK1078 pKa = 11.19NVYY1081 pKa = 9.26QEE1083 pKa = 4.68IEE1085 pKa = 4.44DD1086 pKa = 3.72NTLVRR1091 pKa = 11.84GAIIPKK1097 pKa = 10.31NITTLSWSNVQRR1109 pKa = 11.84YY1110 pKa = 7.47FRR1112 pKa = 11.84FDD1114 pKa = 3.18VYY1116 pKa = 11.2KK1117 pKa = 10.96DD1118 pKa = 4.66DD1119 pKa = 3.93ITDD1122 pKa = 4.09DD1123 pKa = 3.98LTTGTIDD1130 pKa = 3.05TGTGAKK1136 pKa = 8.98MYY1138 pKa = 9.81NFKK1141 pKa = 10.75IIDD1144 pKa = 3.85YY1145 pKa = 10.22QSAPLPFVTQQSGSLPTAVDD1165 pKa = 3.23IASDD1169 pKa = 4.03GVNGADD1175 pKa = 4.43AITYY1179 pKa = 7.95DD1180 pKa = 3.25WSIVRR1185 pKa = 11.84VEE1187 pKa = 4.82HH1188 pKa = 7.94DD1189 pKa = 4.66DD1190 pKa = 2.88ITYY1193 pKa = 10.77NDD1195 pKa = 3.92RR1196 pKa = 11.84QKK1198 pKa = 11.29HH1199 pKa = 5.45LGLFVNEE1206 pKa = 4.69LDD1208 pKa = 4.37AGLNPIEE1215 pKa = 5.24FSVQNDD1221 pKa = 3.58SEE1223 pKa = 5.83LNVSWYY1229 pKa = 10.86LKK1231 pKa = 10.71LDD1233 pKa = 3.44GLIDD1237 pKa = 4.62LEE1239 pKa = 4.89GEE1241 pKa = 4.26SQLVQGSDD1249 pKa = 3.08STLDD1253 pKa = 3.37INSKK1257 pKa = 8.7GTIEE1261 pKa = 4.9RR1262 pKa = 11.84DD1263 pKa = 3.49QQGTADD1269 pKa = 4.48LYY1271 pKa = 10.78TYY1273 pKa = 9.98NYY1275 pKa = 7.23WAAPVGVTSTISNNNNYY1292 pKa = 7.81TLPDD1296 pKa = 3.65VLNDD1300 pKa = 3.52GTTPAIPSSINWLTSGYY1317 pKa = 10.23DD1318 pKa = 3.55GTSGSPVGIADD1329 pKa = 3.37YY1330 pKa = 9.81WIWKK1334 pKa = 7.23YY1335 pKa = 11.44ANRR1338 pKa = 11.84VSDD1341 pKa = 5.27NYY1343 pKa = 10.66PSWQHH1348 pKa = 4.87VRR1350 pKa = 11.84STGSLQPGEE1359 pKa = 4.69GYY1361 pKa = 8.82TMKK1364 pKa = 10.21GTTNTSGAIFTEE1376 pKa = 3.92QNYY1379 pKa = 8.94VFNGKK1384 pKa = 7.73PHH1386 pKa = 6.59NGDD1389 pKa = 2.72ITLTLSAGNDD1399 pKa = 3.55YY1400 pKa = 11.19LIGNPYY1406 pKa = 10.72ASAIDD1411 pKa = 3.71ANEE1414 pKa = 4.58FILDD1418 pKa = 4.36NISDD1422 pKa = 4.01GAGRR1426 pKa = 11.84AGSNIINGTLYY1437 pKa = 10.28FWEE1440 pKa = 4.83HH1441 pKa = 6.05FASSTHH1447 pKa = 6.26ILSEE1451 pKa = 4.08YY1452 pKa = 8.82QGGYY1456 pKa = 8.85GAYY1459 pKa = 9.19TLITGTAAINNDD1471 pKa = 2.61VRR1473 pKa = 11.84INHH1476 pKa = 6.37TGGLTSSKK1484 pKa = 10.2GAPEE1488 pKa = 4.02RR1489 pKa = 11.84YY1490 pKa = 9.41IPVAQGFFVTADD1502 pKa = 3.18TGGSITFKK1510 pKa = 10.55NSQRR1514 pKa = 11.84TFKK1517 pKa = 10.67TEE1519 pKa = 3.56AADD1522 pKa = 3.42PSVFMRR1528 pKa = 11.84VNDD1531 pKa = 4.21GKK1533 pKa = 11.19SKK1535 pKa = 9.96QKK1537 pKa = 10.3KK1538 pKa = 9.73SSVTTSKK1545 pKa = 7.72TTNDD1549 pKa = 2.84TRR1551 pKa = 11.84EE1552 pKa = 4.07VIKK1555 pKa = 10.78LYY1557 pKa = 10.53FDD1559 pKa = 3.63SPKK1562 pKa = 10.54GYY1564 pKa = 9.74HH1565 pKa = 5.53RR1566 pKa = 11.84QIAVGIDD1573 pKa = 3.17SNTTDD1578 pKa = 3.4GHH1580 pKa = 6.88DD1581 pKa = 3.27IGYY1584 pKa = 7.99DD1585 pKa = 3.21APLIEE1590 pKa = 5.95DD1591 pKa = 3.63NVEE1594 pKa = 3.9DD1595 pKa = 4.48MFWTFDD1601 pKa = 3.36NNKK1604 pKa = 9.42FVIQGVSDD1612 pKa = 3.71FNINRR1617 pKa = 11.84TLPLGVKK1624 pKa = 9.32IDD1626 pKa = 3.81QEE1628 pKa = 4.39GTATIRR1634 pKa = 11.84VDD1636 pKa = 3.52YY1637 pKa = 10.8LEE1639 pKa = 5.54NIPIDD1644 pKa = 3.75TNIYY1648 pKa = 10.54LLDD1651 pKa = 4.35KK1652 pKa = 10.47EE1653 pKa = 4.5LEE1655 pKa = 4.17IYY1657 pKa = 10.53HH1658 pKa = 6.96DD1659 pKa = 3.99LRR1661 pKa = 11.84EE1662 pKa = 4.29SNYY1665 pKa = 10.41EE1666 pKa = 3.78VFLTIGEE1673 pKa = 4.27HH1674 pKa = 5.44YY1675 pKa = 10.79DD1676 pKa = 3.14RR1677 pKa = 11.84FEE1679 pKa = 3.87ITFSNGEE1686 pKa = 4.14EE1687 pKa = 3.93EE1688 pKa = 4.11VDD1690 pKa = 4.39PIEE1693 pKa = 5.42GLTKK1697 pKa = 10.54TNLHH1701 pKa = 6.57FSNDD1705 pKa = 3.35KK1706 pKa = 10.77EE1707 pKa = 4.56SIIVINPDD1715 pKa = 2.98NLIINSVKK1723 pKa = 9.59MVNILGQSVFKK1734 pKa = 10.35MDD1736 pKa = 3.2TAFEE1740 pKa = 4.12DD1741 pKa = 4.21RR1742 pKa = 11.84YY1743 pKa = 11.16KK1744 pKa = 10.88EE1745 pKa = 3.76ITVKK1749 pKa = 10.55HH1750 pKa = 6.4LNTGAYY1756 pKa = 8.64IINMITNKK1764 pKa = 10.44GLITKK1769 pKa = 9.9KK1770 pKa = 10.7VVVEE1774 pKa = 4.07
Molecular weight: 192.25 kDa
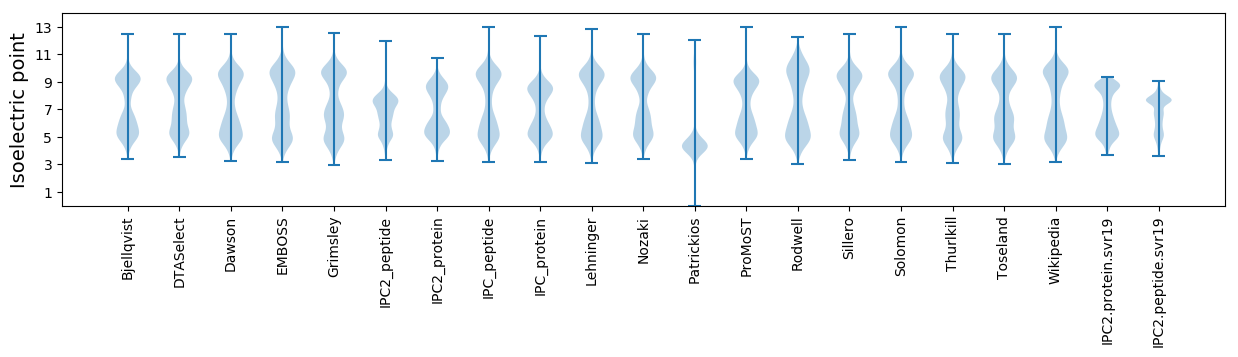
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4V5LQD5|A0A4V5LQD5_9FLAO DUF2807 domain-containing protein OS=Lacinutrix sp. CAU 1491 OX=2565367 GN=E5167_10865 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.27RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.66LSVSSEE47 pKa = 3.9LRR49 pKa = 11.84HH50 pKa = 6.16KK51 pKa = 10.53KK52 pKa = 10.04
MM1 pKa = 7.45KK2 pKa = 9.59RR3 pKa = 11.84TFQPSKK9 pKa = 9.13RR10 pKa = 11.84KK11 pKa = 9.48RR12 pKa = 11.84RR13 pKa = 11.84NKK15 pKa = 9.49HH16 pKa = 3.94GFRR19 pKa = 11.84EE20 pKa = 4.27RR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 10.09GRR39 pKa = 11.84KK40 pKa = 7.97KK41 pKa = 10.66LSVSSEE47 pKa = 3.9LRR49 pKa = 11.84HH50 pKa = 6.16KK51 pKa = 10.53KK52 pKa = 10.04
Molecular weight: 6.25 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
998528 |
28 |
2381 |
334.4 |
37.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.024 ± 0.048 | 0.719 ± 0.013 |
5.624 ± 0.035 | 6.421 ± 0.044 |
5.207 ± 0.035 | 6.132 ± 0.046 |
1.817 ± 0.018 | 8.214 ± 0.045 |
8.226 ± 0.053 | 9.215 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.19 ± 0.023 | 6.684 ± 0.049 |
3.261 ± 0.022 | 3.255 ± 0.023 |
3.32 ± 0.029 | 6.555 ± 0.037 |
5.799 ± 0.042 | 6.145 ± 0.031 |
1.049 ± 0.016 | 4.14 ± 0.031 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
