
Gyrovirus 10
Taxonomy: Viruses; Anelloviridae; Gyrovirus; unclassified Gyrovirus
Average proteome isoelectric point is 8.17
Get precalculated fractions of proteins

Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
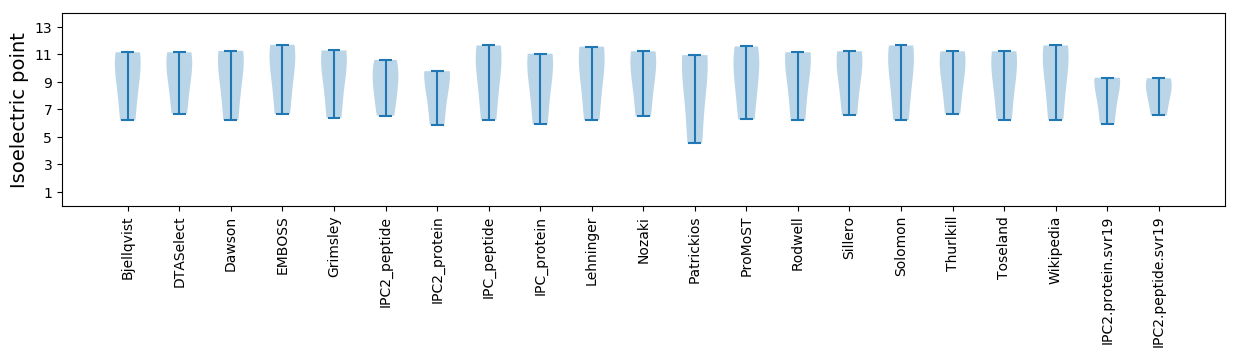
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A2U9N4H7|A0A2U9N4H7_9VIRU Apoptin OS=Gyrovirus 10 OX=2218660 GN=VP3 PE=3 SV=1
MM1 pKa = 7.27AHH3 pKa = 7.45WSPATPIKK11 pKa = 10.45SPPITPPDD19 pKa = 3.15KK20 pKa = 10.66RR21 pKa = 11.84NYY23 pKa = 6.71VTKK26 pKa = 10.15TINGINASNKK36 pKa = 8.36FCNVGWDD43 pKa = 3.65KK44 pKa = 11.25LQNSPDD50 pKa = 3.5WYY52 pKa = 8.75RR53 pKa = 11.84ANYY56 pKa = 9.24NRR58 pKa = 11.84TIATWLRR65 pKa = 11.84DD66 pKa = 3.5CKK68 pKa = 9.82TAHH71 pKa = 6.76SKK73 pKa = 9.81ICSCSSFRR81 pKa = 11.84NHH83 pKa = 5.41WHH85 pKa = 5.23QTDD88 pKa = 3.33ATLTTDD94 pKa = 3.99SSTQTDD100 pKa = 3.52VDD102 pKa = 3.55TSPIIRR108 pKa = 11.84AGEE111 pKa = 3.75RR112 pKa = 11.84ARR114 pKa = 11.84RR115 pKa = 11.84KK116 pKa = 9.05ILARR120 pKa = 11.84EE121 pKa = 3.95RR122 pKa = 11.84RR123 pKa = 11.84QATPGNRR130 pKa = 11.84KK131 pKa = 9.21KK132 pKa = 10.46KK133 pKa = 9.79HH134 pKa = 5.55KK135 pKa = 9.32TVTWDD140 pKa = 3.0IGPEE144 pKa = 4.0EE145 pKa = 4.74SSTPSDD151 pKa = 3.53EE152 pKa = 5.8DD153 pKa = 3.72DD154 pKa = 4.72GEE156 pKa = 5.0SIDD159 pKa = 5.83AFDD162 pKa = 4.73LTEE165 pKa = 6.02DD166 pKa = 4.68DD167 pKa = 5.34DD168 pKa = 6.25DD169 pKa = 4.55SAFANVNFDD178 pKa = 4.23LATDD182 pKa = 4.3LDD184 pKa = 4.17AALNAPSGTNTPARR198 pKa = 11.84MWW200 pKa = 3.83
MM1 pKa = 7.27AHH3 pKa = 7.45WSPATPIKK11 pKa = 10.45SPPITPPDD19 pKa = 3.15KK20 pKa = 10.66RR21 pKa = 11.84NYY23 pKa = 6.71VTKK26 pKa = 10.15TINGINASNKK36 pKa = 8.36FCNVGWDD43 pKa = 3.65KK44 pKa = 11.25LQNSPDD50 pKa = 3.5WYY52 pKa = 8.75RR53 pKa = 11.84ANYY56 pKa = 9.24NRR58 pKa = 11.84TIATWLRR65 pKa = 11.84DD66 pKa = 3.5CKK68 pKa = 9.82TAHH71 pKa = 6.76SKK73 pKa = 9.81ICSCSSFRR81 pKa = 11.84NHH83 pKa = 5.41WHH85 pKa = 5.23QTDD88 pKa = 3.33ATLTTDD94 pKa = 3.99SSTQTDD100 pKa = 3.52VDD102 pKa = 3.55TSPIIRR108 pKa = 11.84AGEE111 pKa = 3.75RR112 pKa = 11.84ARR114 pKa = 11.84RR115 pKa = 11.84KK116 pKa = 9.05ILARR120 pKa = 11.84EE121 pKa = 3.95RR122 pKa = 11.84RR123 pKa = 11.84QATPGNRR130 pKa = 11.84KK131 pKa = 9.21KK132 pKa = 10.46KK133 pKa = 9.79HH134 pKa = 5.55KK135 pKa = 9.32TVTWDD140 pKa = 3.0IGPEE144 pKa = 4.0EE145 pKa = 4.74SSTPSDD151 pKa = 3.53EE152 pKa = 5.8DD153 pKa = 3.72DD154 pKa = 4.72GEE156 pKa = 5.0SIDD159 pKa = 5.83AFDD162 pKa = 4.73LTEE165 pKa = 6.02DD166 pKa = 4.68DD167 pKa = 5.34DD168 pKa = 6.25DD169 pKa = 4.55SAFANVNFDD178 pKa = 4.23LATDD182 pKa = 4.3LDD184 pKa = 4.17AALNAPSGTNTPARR198 pKa = 11.84MWW200 pKa = 3.83
Molecular weight: 22.43 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2U9N6R0|A0A2U9N6R0_9VIRU CA1 OS=Gyrovirus 10 OX=2218660 GN=VP1 PE=3 SV=1
MM1 pKa = 7.98GYY3 pKa = 10.06RR4 pKa = 11.84SRR6 pKa = 11.84GKK8 pKa = 9.6FYY10 pKa = 9.87TFRR13 pKa = 11.84RR14 pKa = 11.84GRR16 pKa = 11.84WRR18 pKa = 11.84KK19 pKa = 8.41HH20 pKa = 3.91RR21 pKa = 11.84RR22 pKa = 11.84FRR24 pKa = 11.84PYY26 pKa = 9.25RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84FRR33 pKa = 11.84FRR35 pKa = 11.84KK36 pKa = 8.53RR37 pKa = 11.84KK38 pKa = 8.72FRR40 pKa = 11.84PRR42 pKa = 11.84YY43 pKa = 7.73GFRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84FKK50 pKa = 9.75RR51 pKa = 11.84AFWNQHH57 pKa = 4.93PGSYY61 pKa = 9.62VVRR64 pKa = 11.84LPNPQNTLNLMFQGIVWFPLPKK86 pKa = 10.37YY87 pKa = 10.13CINSTNTLKK96 pKa = 11.12NIDD99 pKa = 3.82VQNVITCRR107 pKa = 11.84VGSIHH112 pKa = 7.44ISLKK116 pKa = 10.19QFLLAVMSLSGTTKK130 pKa = 10.74LGGPSIGKK138 pKa = 9.82LYY140 pKa = 11.22YY141 pKa = 9.56EE142 pKa = 5.04AKK144 pKa = 10.43DD145 pKa = 3.55KK146 pKa = 11.25LGQAGNYY153 pKa = 9.89PYY155 pKa = 9.45MEE157 pKa = 5.51KK158 pKa = 10.01IASQTQSASLPSQWWRR174 pKa = 11.84WAIMFMYY181 pKa = 10.31PPDD184 pKa = 3.99NKK186 pKa = 10.9LIDD189 pKa = 4.14YY190 pKa = 9.28FPAQPDD196 pKa = 3.79PKK198 pKa = 10.52DD199 pKa = 3.51LQNILGGWSLFRR211 pKa = 11.84HH212 pKa = 6.18KK213 pKa = 9.2KK214 pKa = 7.72TKK216 pKa = 9.84VAIIATSPQGNWSPVASLTSQDD238 pKa = 3.58SYY240 pKa = 11.93FMRR243 pKa = 11.84HH244 pKa = 5.97CSNVFAVTKK253 pKa = 10.83GNFPLSGAVGTKK265 pKa = 9.48TPTSFLPDD273 pKa = 3.04EE274 pKa = 4.87CEE276 pKa = 4.18YY277 pKa = 10.6PSRR280 pKa = 11.84PPQAEE285 pKa = 3.88QTNQTGIPGQCYY297 pKa = 8.21TRR299 pKa = 11.84QPQDD303 pKa = 3.03GLVEE307 pKa = 4.16TDD309 pKa = 2.69CWYY312 pKa = 10.53SQEE315 pKa = 4.3TFPSFGTLSALGASWAFPPGQRR337 pKa = 11.84SFSHH341 pKa = 6.63GSVNHH346 pKa = 5.03HH347 pKa = 7.26TITGSGDD354 pKa = 3.32PEE356 pKa = 4.07GQRR359 pKa = 11.84WRR361 pKa = 11.84TLNPLTTFPITSSEE375 pKa = 3.95QGSDD379 pKa = 3.42YY380 pKa = 11.41DD381 pKa = 3.96RR382 pKa = 11.84VIASIYY388 pKa = 9.37LAQGSDD394 pKa = 2.75QFLPYY399 pKa = 10.31KK400 pKa = 10.66YY401 pKa = 9.96GTDD404 pKa = 3.13QDD406 pKa = 3.86QPNWRR411 pKa = 11.84PQKK414 pKa = 9.3FTQQMNVWARR424 pKa = 11.84IGEE427 pKa = 4.14FRR429 pKa = 11.84AA430 pKa = 3.62
MM1 pKa = 7.98GYY3 pKa = 10.06RR4 pKa = 11.84SRR6 pKa = 11.84GKK8 pKa = 9.6FYY10 pKa = 9.87TFRR13 pKa = 11.84RR14 pKa = 11.84GRR16 pKa = 11.84WRR18 pKa = 11.84KK19 pKa = 8.41HH20 pKa = 3.91RR21 pKa = 11.84RR22 pKa = 11.84FRR24 pKa = 11.84PYY26 pKa = 9.25RR27 pKa = 11.84RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84RR31 pKa = 11.84FRR33 pKa = 11.84FRR35 pKa = 11.84KK36 pKa = 8.53RR37 pKa = 11.84KK38 pKa = 8.72FRR40 pKa = 11.84PRR42 pKa = 11.84YY43 pKa = 7.73GFRR46 pKa = 11.84RR47 pKa = 11.84RR48 pKa = 11.84FKK50 pKa = 9.75RR51 pKa = 11.84AFWNQHH57 pKa = 4.93PGSYY61 pKa = 9.62VVRR64 pKa = 11.84LPNPQNTLNLMFQGIVWFPLPKK86 pKa = 10.37YY87 pKa = 10.13CINSTNTLKK96 pKa = 11.12NIDD99 pKa = 3.82VQNVITCRR107 pKa = 11.84VGSIHH112 pKa = 7.44ISLKK116 pKa = 10.19QFLLAVMSLSGTTKK130 pKa = 10.74LGGPSIGKK138 pKa = 9.82LYY140 pKa = 11.22YY141 pKa = 9.56EE142 pKa = 5.04AKK144 pKa = 10.43DD145 pKa = 3.55KK146 pKa = 11.25LGQAGNYY153 pKa = 9.89PYY155 pKa = 9.45MEE157 pKa = 5.51KK158 pKa = 10.01IASQTQSASLPSQWWRR174 pKa = 11.84WAIMFMYY181 pKa = 10.31PPDD184 pKa = 3.99NKK186 pKa = 10.9LIDD189 pKa = 4.14YY190 pKa = 9.28FPAQPDD196 pKa = 3.79PKK198 pKa = 10.52DD199 pKa = 3.51LQNILGGWSLFRR211 pKa = 11.84HH212 pKa = 6.18KK213 pKa = 9.2KK214 pKa = 7.72TKK216 pKa = 9.84VAIIATSPQGNWSPVASLTSQDD238 pKa = 3.58SYY240 pKa = 11.93FMRR243 pKa = 11.84HH244 pKa = 5.97CSNVFAVTKK253 pKa = 10.83GNFPLSGAVGTKK265 pKa = 9.48TPTSFLPDD273 pKa = 3.04EE274 pKa = 4.87CEE276 pKa = 4.18YY277 pKa = 10.6PSRR280 pKa = 11.84PPQAEE285 pKa = 3.88QTNQTGIPGQCYY297 pKa = 8.21TRR299 pKa = 11.84QPQDD303 pKa = 3.03GLVEE307 pKa = 4.16TDD309 pKa = 2.69CWYY312 pKa = 10.53SQEE315 pKa = 4.3TFPSFGTLSALGASWAFPPGQRR337 pKa = 11.84SFSHH341 pKa = 6.63GSVNHH346 pKa = 5.03HH347 pKa = 7.26TITGSGDD354 pKa = 3.32PEE356 pKa = 4.07GQRR359 pKa = 11.84WRR361 pKa = 11.84TLNPLTTFPITSSEE375 pKa = 3.95QGSDD379 pKa = 3.42YY380 pKa = 11.41DD381 pKa = 3.96RR382 pKa = 11.84VIASIYY388 pKa = 9.37LAQGSDD394 pKa = 2.75QFLPYY399 pKa = 10.31KK400 pKa = 10.66YY401 pKa = 9.96GTDD404 pKa = 3.13QDD406 pKa = 3.86QPNWRR411 pKa = 11.84PQKK414 pKa = 9.3FTQQMNVWARR424 pKa = 11.84IGEE427 pKa = 4.14FRR429 pKa = 11.84AA430 pKa = 3.62
Molecular weight: 49.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
738 |
108 |
430 |
246.0 |
27.97 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.962 ± 1.434 | 1.491 ± 0.208 |
5.149 ± 2.154 | 2.981 ± 0.56 |
4.336 ± 1.64 | 6.504 ± 1.273 |
2.304 ± 0.398 | 4.878 ± 0.389 |
5.014 ± 0.531 | 6.233 ± 1.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.762 ± 0.321 | 4.743 ± 0.943 |
7.588 ± 0.44 | 5.556 ± 1.519 |
8.266 ± 0.51 | 8.672 ± 0.577 |
8.672 ± 1.622 | 3.794 ± 0.657 |
2.846 ± 0.432 | 3.252 ± 1.179 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
