
Gammapapillomavirus 12
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
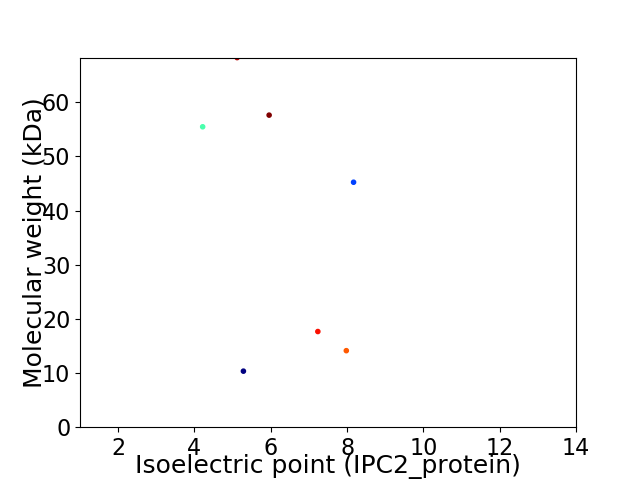
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A2D2ALT7|A0A2D2ALT7_9PAPI ^E4 (Fragment) OS=Gammapapillomavirus 12 OX=1513257 GN=^E4 PE=4 SV=1
MM1 pKa = 7.34SGSIRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.18RR9 pKa = 11.84ASPTDD14 pKa = 3.76LYY16 pKa = 10.49KK17 pKa = 11.02ACVQGADD24 pKa = 4.46CISDD28 pKa = 3.46VKK30 pKa = 11.31NKK32 pKa = 10.16FEE34 pKa = 4.03QTTIADD40 pKa = 3.73WLLKK44 pKa = 10.4IFGSLVYY51 pKa = 10.34FGNLGIGTGKK61 pKa = 10.78GSGGSLGFRR70 pKa = 11.84PLGEE74 pKa = 4.0GVGAGKK80 pKa = 9.56PITTGAIKK88 pKa = 9.89PAVAVEE94 pKa = 3.94TLPVDD99 pKa = 3.78VFAIPSEE106 pKa = 3.95ASAIVPLAEE115 pKa = 5.05GGPDD119 pKa = 3.33LNVFTTDD126 pKa = 3.32GGPGLGAEE134 pKa = 4.46EE135 pKa = 4.39IEE137 pKa = 5.26LYY139 pKa = 10.41TITDD143 pKa = 3.35QTVDD147 pKa = 3.22GDD149 pKa = 4.7SIGGNPNIVTNEE161 pKa = 3.83RR162 pKa = 11.84GVPAVLDD169 pKa = 3.71VQPTPEE175 pKa = 4.65RR176 pKa = 11.84PPQVLYY182 pKa = 11.03DD183 pKa = 3.64PTIDD187 pKa = 2.95ATAQIRR193 pKa = 11.84IVTSDD198 pKa = 3.33PTLTQDD204 pKa = 3.11TNIFVDD210 pKa = 4.58PYY212 pKa = 10.64VGGEE216 pKa = 3.88IVGTGSRR223 pKa = 11.84FEE225 pKa = 4.89EE226 pKa = 4.28IPLDD230 pKa = 4.04EE231 pKa = 4.34FTLGQFEE238 pKa = 4.61IEE240 pKa = 4.18EE241 pKa = 4.46SPITSTPTEE250 pKa = 3.99RR251 pKa = 11.84LDD253 pKa = 3.46NAVNRR258 pKa = 11.84VRR260 pKa = 11.84GFYY263 pKa = 10.54NRR265 pKa = 11.84LIRR268 pKa = 11.84QVPVTDD274 pKa = 4.15PVFLQQPSRR283 pKa = 11.84LVQFEE288 pKa = 4.47YY289 pKa = 10.88EE290 pKa = 4.43NPAFDD295 pKa = 5.75DD296 pKa = 4.2DD297 pKa = 4.34VSLTFEE303 pKa = 4.65RR304 pKa = 11.84DD305 pKa = 3.12LAAVTAAPHH314 pKa = 6.63EE315 pKa = 4.57DD316 pKa = 3.18FADD319 pKa = 3.38IVKK322 pKa = 10.67LSRR325 pKa = 11.84PEE327 pKa = 3.94LSEE330 pKa = 4.05SVPGVVRR337 pKa = 11.84VSRR340 pKa = 11.84LGEE343 pKa = 4.12TNTITTRR350 pKa = 11.84SGTTIGQKK358 pKa = 7.9VHH360 pKa = 6.95FYY362 pKa = 11.02YY363 pKa = 10.68DD364 pKa = 2.71ISTIQEE370 pKa = 3.87AEE372 pKa = 4.13NIEE375 pKa = 4.23LQTLGEE381 pKa = 4.27HH382 pKa = 6.45TGTSTIVDD390 pKa = 5.02DD391 pKa = 4.55ILASTIVDD399 pKa = 3.66PVNNADD405 pKa = 3.85VGISEE410 pKa = 5.81DD411 pKa = 3.63NLLDD415 pKa = 3.62TFAEE419 pKa = 4.49DD420 pKa = 3.74FNNAHH425 pKa = 6.9LVLQAAGNEE434 pKa = 4.17DD435 pKa = 4.04EE436 pKa = 6.0DD437 pKa = 4.02IVSIPLLPPGAALKK451 pKa = 10.92LFVADD456 pKa = 3.66VGDD459 pKa = 4.17GLFVSYY465 pKa = 8.44PTDD468 pKa = 3.82TIHH471 pKa = 8.08DD472 pKa = 3.79ITIQLPDD479 pKa = 4.38GLPIGPSYY487 pKa = 10.94YY488 pKa = 10.8VGVDD492 pKa = 2.73NDD494 pKa = 4.44FYY496 pKa = 11.31LHH498 pKa = 7.23PSLIPKK504 pKa = 9.77KK505 pKa = 9.65KK506 pKa = 9.47RR507 pKa = 11.84RR508 pKa = 11.84RR509 pKa = 11.84LEE511 pKa = 3.84YY512 pKa = 10.83YY513 pKa = 10.02FF514 pKa = 5.21
MM1 pKa = 7.34SGSIRR6 pKa = 11.84RR7 pKa = 11.84KK8 pKa = 9.18RR9 pKa = 11.84ASPTDD14 pKa = 3.76LYY16 pKa = 10.49KK17 pKa = 11.02ACVQGADD24 pKa = 4.46CISDD28 pKa = 3.46VKK30 pKa = 11.31NKK32 pKa = 10.16FEE34 pKa = 4.03QTTIADD40 pKa = 3.73WLLKK44 pKa = 10.4IFGSLVYY51 pKa = 10.34FGNLGIGTGKK61 pKa = 10.78GSGGSLGFRR70 pKa = 11.84PLGEE74 pKa = 4.0GVGAGKK80 pKa = 9.56PITTGAIKK88 pKa = 9.89PAVAVEE94 pKa = 3.94TLPVDD99 pKa = 3.78VFAIPSEE106 pKa = 3.95ASAIVPLAEE115 pKa = 5.05GGPDD119 pKa = 3.33LNVFTTDD126 pKa = 3.32GGPGLGAEE134 pKa = 4.46EE135 pKa = 4.39IEE137 pKa = 5.26LYY139 pKa = 10.41TITDD143 pKa = 3.35QTVDD147 pKa = 3.22GDD149 pKa = 4.7SIGGNPNIVTNEE161 pKa = 3.83RR162 pKa = 11.84GVPAVLDD169 pKa = 3.71VQPTPEE175 pKa = 4.65RR176 pKa = 11.84PPQVLYY182 pKa = 11.03DD183 pKa = 3.64PTIDD187 pKa = 2.95ATAQIRR193 pKa = 11.84IVTSDD198 pKa = 3.33PTLTQDD204 pKa = 3.11TNIFVDD210 pKa = 4.58PYY212 pKa = 10.64VGGEE216 pKa = 3.88IVGTGSRR223 pKa = 11.84FEE225 pKa = 4.89EE226 pKa = 4.28IPLDD230 pKa = 4.04EE231 pKa = 4.34FTLGQFEE238 pKa = 4.61IEE240 pKa = 4.18EE241 pKa = 4.46SPITSTPTEE250 pKa = 3.99RR251 pKa = 11.84LDD253 pKa = 3.46NAVNRR258 pKa = 11.84VRR260 pKa = 11.84GFYY263 pKa = 10.54NRR265 pKa = 11.84LIRR268 pKa = 11.84QVPVTDD274 pKa = 4.15PVFLQQPSRR283 pKa = 11.84LVQFEE288 pKa = 4.47YY289 pKa = 10.88EE290 pKa = 4.43NPAFDD295 pKa = 5.75DD296 pKa = 4.2DD297 pKa = 4.34VSLTFEE303 pKa = 4.65RR304 pKa = 11.84DD305 pKa = 3.12LAAVTAAPHH314 pKa = 6.63EE315 pKa = 4.57DD316 pKa = 3.18FADD319 pKa = 3.38IVKK322 pKa = 10.67LSRR325 pKa = 11.84PEE327 pKa = 3.94LSEE330 pKa = 4.05SVPGVVRR337 pKa = 11.84VSRR340 pKa = 11.84LGEE343 pKa = 4.12TNTITTRR350 pKa = 11.84SGTTIGQKK358 pKa = 7.9VHH360 pKa = 6.95FYY362 pKa = 11.02YY363 pKa = 10.68DD364 pKa = 2.71ISTIQEE370 pKa = 3.87AEE372 pKa = 4.13NIEE375 pKa = 4.23LQTLGEE381 pKa = 4.27HH382 pKa = 6.45TGTSTIVDD390 pKa = 5.02DD391 pKa = 4.55ILASTIVDD399 pKa = 3.66PVNNADD405 pKa = 3.85VGISEE410 pKa = 5.81DD411 pKa = 3.63NLLDD415 pKa = 3.62TFAEE419 pKa = 4.49DD420 pKa = 3.74FNNAHH425 pKa = 6.9LVLQAAGNEE434 pKa = 4.17DD435 pKa = 4.04EE436 pKa = 6.0DD437 pKa = 4.02IVSIPLLPPGAALKK451 pKa = 10.92LFVADD456 pKa = 3.66VGDD459 pKa = 4.17GLFVSYY465 pKa = 8.44PTDD468 pKa = 3.82TIHH471 pKa = 8.08DD472 pKa = 3.79ITIQLPDD479 pKa = 4.38GLPIGPSYY487 pKa = 10.94YY488 pKa = 10.8VGVDD492 pKa = 2.73NDD494 pKa = 4.44FYY496 pKa = 11.31LHH498 pKa = 7.23PSLIPKK504 pKa = 9.77KK505 pKa = 9.65KK506 pKa = 9.47RR507 pKa = 11.84RR508 pKa = 11.84RR509 pKa = 11.84LEE511 pKa = 3.84YY512 pKa = 10.83YY513 pKa = 10.02FF514 pKa = 5.21
Molecular weight: 55.49 kDa
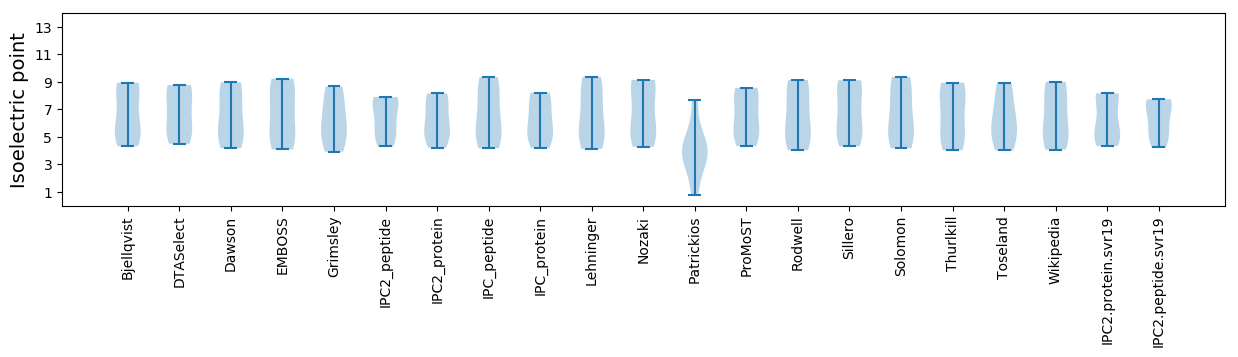
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2D2ALT7|A0A2D2ALT7_9PAPI ^E4 (Fragment) OS=Gammapapillomavirus 12 OX=1513257 GN=^E4 PE=4 SV=1
SS1 pKa = 7.02IKK3 pKa = 10.3IKK5 pKa = 10.84LFLPLCLASQGLFPSLRR22 pKa = 11.84RR23 pKa = 11.84SLSPPPTPHH32 pKa = 6.9PHH34 pKa = 6.16RR35 pKa = 11.84KK36 pKa = 8.54SLEE39 pKa = 4.08RR40 pKa = 11.84EE41 pKa = 4.12GEE43 pKa = 4.09KK44 pKa = 10.19PRR46 pKa = 11.84RR47 pKa = 11.84TPTPNRR53 pKa = 11.84PPRR56 pKa = 11.84PTLNFDD62 pKa = 4.51EE63 pKa = 6.06DD64 pKa = 4.17DD65 pKa = 4.15EE66 pKa = 4.74NNKK69 pKa = 10.02EE70 pKa = 4.01NLPPPEE76 pKa = 4.3YY77 pKa = 10.59EE78 pKa = 4.31EE79 pKa = 4.26EE80 pKa = 4.17QQGAATTLHH89 pKa = 5.28QLLRR93 pKa = 11.84KK94 pKa = 9.48WEE96 pKa = 4.05VEE98 pKa = 3.4LDD100 pKa = 3.42RR101 pKa = 11.84FRR103 pKa = 11.84DD104 pKa = 3.85TVIQDD109 pKa = 3.48LDD111 pKa = 3.65AFKK114 pKa = 10.88KK115 pKa = 10.56RR116 pKa = 11.84LGIRR120 pKa = 11.84HH121 pKa = 6.09
SS1 pKa = 7.02IKK3 pKa = 10.3IKK5 pKa = 10.84LFLPLCLASQGLFPSLRR22 pKa = 11.84RR23 pKa = 11.84SLSPPPTPHH32 pKa = 6.9PHH34 pKa = 6.16RR35 pKa = 11.84KK36 pKa = 8.54SLEE39 pKa = 4.08RR40 pKa = 11.84EE41 pKa = 4.12GEE43 pKa = 4.09KK44 pKa = 10.19PRR46 pKa = 11.84RR47 pKa = 11.84TPTPNRR53 pKa = 11.84PPRR56 pKa = 11.84PTLNFDD62 pKa = 4.51EE63 pKa = 6.06DD64 pKa = 4.17DD65 pKa = 4.15EE66 pKa = 4.74NNKK69 pKa = 10.02EE70 pKa = 4.01NLPPPEE76 pKa = 4.3YY77 pKa = 10.59EE78 pKa = 4.31EE79 pKa = 4.26EE80 pKa = 4.17QQGAATTLHH89 pKa = 5.28QLLRR93 pKa = 11.84KK94 pKa = 9.48WEE96 pKa = 4.05VEE98 pKa = 3.4LDD100 pKa = 3.42RR101 pKa = 11.84FRR103 pKa = 11.84DD104 pKa = 3.85TVIQDD109 pKa = 3.48LDD111 pKa = 3.65AFKK114 pKa = 10.88KK115 pKa = 10.56RR116 pKa = 11.84LGIRR120 pKa = 11.84HH121 pKa = 6.09
Molecular weight: 14.14 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2379 |
92 |
599 |
339.9 |
38.4 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.557 ± 0.612 | 2.522 ± 0.956 |
6.389 ± 0.67 | 6.81 ± 0.51 |
4.75 ± 0.47 | 5.591 ± 1.194 |
1.765 ± 0.31 | 5.002 ± 0.718 |
5.633 ± 0.921 | 9.332 ± 0.45 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.429 ± 0.47 | 5.212 ± 0.522 |
5.717 ± 1.012 | 3.993 ± 0.23 |
5.591 ± 0.678 | 6.726 ± 0.477 |
6.011 ± 0.692 | 6.053 ± 0.73 |
1.303 ± 0.293 | 3.615 ± 0.35 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
