
Changjiang astro-like virus
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified RNA viruses ShiM-2016
Average proteome isoelectric point is 6.87
Get precalculated fractions of proteins
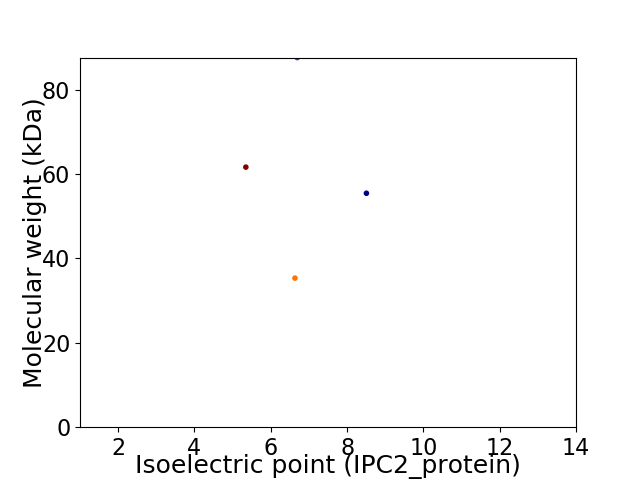
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1L3KNS4|A0A1L3KNS4_9VIRU Capsid protein OS=Changjiang astro-like virus OX=1922764 PE=3 SV=1
MM1 pKa = 7.88GGMRR5 pKa = 11.84WQMRR9 pKa = 11.84EE10 pKa = 3.99NTPTLQDD17 pKa = 3.33NTPIWEE23 pKa = 3.96RR24 pKa = 11.84WEE26 pKa = 4.43EE27 pKa = 3.44IDD29 pKa = 5.31YY30 pKa = 10.19PPEE33 pKa = 4.34PEE35 pKa = 3.73WKK37 pKa = 10.13GRR39 pKa = 11.84SLLMEE44 pKa = 4.7DD45 pKa = 3.03VHH47 pKa = 8.77ALEE50 pKa = 4.1NHH52 pKa = 6.05GMPVYY57 pKa = 9.18NTSFSSFTVVANVNINRR74 pKa = 11.84PCSKK78 pKa = 10.3KK79 pKa = 10.6DD80 pKa = 3.05PAPNVEE86 pKa = 4.4CEE88 pKa = 4.07MAVMSVLEE96 pKa = 4.42GYY98 pKa = 9.0RR99 pKa = 11.84AKK101 pKa = 10.68LSEE104 pKa = 4.14YY105 pKa = 10.07QRR107 pKa = 11.84SGWDD111 pKa = 3.23MEE113 pKa = 4.38VLEE116 pKa = 4.2NTFNKK121 pKa = 9.98FKK123 pKa = 9.22THH125 pKa = 5.49VYY127 pKa = 10.19NVNQCAWTAASDD139 pKa = 4.06VMRR142 pKa = 11.84HH143 pKa = 5.46EE144 pKa = 4.05YY145 pKa = 10.31RR146 pKa = 11.84YY147 pKa = 8.76VPRR150 pKa = 11.84RR151 pKa = 11.84IVDD154 pKa = 3.64FHH156 pKa = 6.66EE157 pKa = 4.2IKK159 pKa = 10.84KK160 pKa = 10.2PVDD163 pKa = 3.94KK164 pKa = 11.3SPGWPGINFASTIEE178 pKa = 4.1EE179 pKa = 3.89WRR181 pKa = 11.84EE182 pKa = 3.97LNPHH186 pKa = 6.37RR187 pKa = 11.84AISLWAEE194 pKa = 3.7MSKK197 pKa = 10.71RR198 pKa = 11.84PVFTGAYY205 pKa = 9.9AFIKK209 pKa = 10.38DD210 pKa = 3.72EE211 pKa = 4.77PIKK214 pKa = 10.75VEE216 pKa = 4.17KK217 pKa = 10.5IKK219 pKa = 10.27TKK221 pKa = 8.61DD222 pKa = 3.18QRR224 pKa = 11.84LILCWDD230 pKa = 4.47DD231 pKa = 5.02SFQLCKK237 pKa = 10.62LRR239 pKa = 11.84FQYY242 pKa = 10.61DD243 pKa = 2.78DD244 pKa = 3.4HH245 pKa = 8.22MIMKK249 pKa = 9.34RR250 pKa = 11.84NWMQSEE256 pKa = 4.52SKK258 pKa = 10.05MGWSPLIGGLHH269 pKa = 7.16DD270 pKa = 5.58SIAPLLSYY278 pKa = 10.12RR279 pKa = 11.84YY280 pKa = 9.89KK281 pKa = 10.6LQEE284 pKa = 3.89DD285 pKa = 4.97FKK287 pKa = 11.08RR288 pKa = 11.84FDD290 pKa = 3.35GTICEE295 pKa = 4.07EE296 pKa = 4.13LMLEE300 pKa = 4.48VYY302 pKa = 11.29SMDD305 pKa = 3.43WDD307 pKa = 3.8NLSYY311 pKa = 10.42EE312 pKa = 4.91HH313 pKa = 6.65KK314 pKa = 9.79TEE316 pKa = 4.54DD317 pKa = 3.82NNMRR321 pKa = 11.84YY322 pKa = 10.25YY323 pKa = 10.97NIVHH327 pKa = 6.2NSIHH331 pKa = 4.83THH333 pKa = 6.42EE334 pKa = 4.84ILPNGDD340 pKa = 3.57VVRR343 pKa = 11.84LAHH346 pKa = 6.28GNKK349 pKa = 9.94SGTSDD354 pKa = 3.52TTPLNAKK361 pKa = 9.93VNTFLKK367 pKa = 9.61AYY369 pKa = 9.7EE370 pKa = 3.96MAAFLLSQGEE380 pKa = 4.34SAEE383 pKa = 4.43EE384 pKa = 3.65ILDD387 pKa = 3.74YY388 pKa = 10.53TYY390 pKa = 11.24HH391 pKa = 7.66DD392 pKa = 3.44WRR394 pKa = 11.84QLYY397 pKa = 8.91TMITYY402 pKa = 10.86GDD404 pKa = 3.86DD405 pKa = 3.46RR406 pKa = 11.84LVGEE410 pKa = 4.83DD411 pKa = 4.38LEE413 pKa = 5.38IPLEE417 pKa = 4.14FKK419 pKa = 10.93EE420 pKa = 4.54NIYY423 pKa = 10.8KK424 pKa = 10.05EE425 pKa = 4.02LGMWLPKK432 pKa = 10.64DD433 pKa = 3.4KK434 pKa = 10.55IKK436 pKa = 10.97RR437 pKa = 11.84QDD439 pKa = 3.55YY440 pKa = 10.51VEE442 pKa = 4.45GLSFCGAVIKK452 pKa = 10.44RR453 pKa = 11.84DD454 pKa = 3.69PCTKK458 pKa = 9.81RR459 pKa = 11.84YY460 pKa = 9.22YY461 pKa = 10.2PCFGDD466 pKa = 3.43NDD468 pKa = 3.9KK469 pKa = 11.56LIDD472 pKa = 3.72SFVWKK477 pKa = 10.4NADD480 pKa = 3.76FEE482 pKa = 4.89EE483 pKa = 4.63KK484 pKa = 10.55LLNYY488 pKa = 9.98VLISYY493 pKa = 8.67GGKK496 pKa = 9.38YY497 pKa = 7.67HH498 pKa = 7.26TSFLKK503 pKa = 8.73MAEE506 pKa = 4.18MNGVHH511 pKa = 6.25VPDD514 pKa = 4.46PQDD517 pKa = 3.03IRR519 pKa = 11.84SLIVGWW525 pKa = 3.7
MM1 pKa = 7.88GGMRR5 pKa = 11.84WQMRR9 pKa = 11.84EE10 pKa = 3.99NTPTLQDD17 pKa = 3.33NTPIWEE23 pKa = 3.96RR24 pKa = 11.84WEE26 pKa = 4.43EE27 pKa = 3.44IDD29 pKa = 5.31YY30 pKa = 10.19PPEE33 pKa = 4.34PEE35 pKa = 3.73WKK37 pKa = 10.13GRR39 pKa = 11.84SLLMEE44 pKa = 4.7DD45 pKa = 3.03VHH47 pKa = 8.77ALEE50 pKa = 4.1NHH52 pKa = 6.05GMPVYY57 pKa = 9.18NTSFSSFTVVANVNINRR74 pKa = 11.84PCSKK78 pKa = 10.3KK79 pKa = 10.6DD80 pKa = 3.05PAPNVEE86 pKa = 4.4CEE88 pKa = 4.07MAVMSVLEE96 pKa = 4.42GYY98 pKa = 9.0RR99 pKa = 11.84AKK101 pKa = 10.68LSEE104 pKa = 4.14YY105 pKa = 10.07QRR107 pKa = 11.84SGWDD111 pKa = 3.23MEE113 pKa = 4.38VLEE116 pKa = 4.2NTFNKK121 pKa = 9.98FKK123 pKa = 9.22THH125 pKa = 5.49VYY127 pKa = 10.19NVNQCAWTAASDD139 pKa = 4.06VMRR142 pKa = 11.84HH143 pKa = 5.46EE144 pKa = 4.05YY145 pKa = 10.31RR146 pKa = 11.84YY147 pKa = 8.76VPRR150 pKa = 11.84RR151 pKa = 11.84IVDD154 pKa = 3.64FHH156 pKa = 6.66EE157 pKa = 4.2IKK159 pKa = 10.84KK160 pKa = 10.2PVDD163 pKa = 3.94KK164 pKa = 11.3SPGWPGINFASTIEE178 pKa = 4.1EE179 pKa = 3.89WRR181 pKa = 11.84EE182 pKa = 3.97LNPHH186 pKa = 6.37RR187 pKa = 11.84AISLWAEE194 pKa = 3.7MSKK197 pKa = 10.71RR198 pKa = 11.84PVFTGAYY205 pKa = 9.9AFIKK209 pKa = 10.38DD210 pKa = 3.72EE211 pKa = 4.77PIKK214 pKa = 10.75VEE216 pKa = 4.17KK217 pKa = 10.5IKK219 pKa = 10.27TKK221 pKa = 8.61DD222 pKa = 3.18QRR224 pKa = 11.84LILCWDD230 pKa = 4.47DD231 pKa = 5.02SFQLCKK237 pKa = 10.62LRR239 pKa = 11.84FQYY242 pKa = 10.61DD243 pKa = 2.78DD244 pKa = 3.4HH245 pKa = 8.22MIMKK249 pKa = 9.34RR250 pKa = 11.84NWMQSEE256 pKa = 4.52SKK258 pKa = 10.05MGWSPLIGGLHH269 pKa = 7.16DD270 pKa = 5.58SIAPLLSYY278 pKa = 10.12RR279 pKa = 11.84YY280 pKa = 9.89KK281 pKa = 10.6LQEE284 pKa = 3.89DD285 pKa = 4.97FKK287 pKa = 11.08RR288 pKa = 11.84FDD290 pKa = 3.35GTICEE295 pKa = 4.07EE296 pKa = 4.13LMLEE300 pKa = 4.48VYY302 pKa = 11.29SMDD305 pKa = 3.43WDD307 pKa = 3.8NLSYY311 pKa = 10.42EE312 pKa = 4.91HH313 pKa = 6.65KK314 pKa = 9.79TEE316 pKa = 4.54DD317 pKa = 3.82NNMRR321 pKa = 11.84YY322 pKa = 10.25YY323 pKa = 10.97NIVHH327 pKa = 6.2NSIHH331 pKa = 4.83THH333 pKa = 6.42EE334 pKa = 4.84ILPNGDD340 pKa = 3.57VVRR343 pKa = 11.84LAHH346 pKa = 6.28GNKK349 pKa = 9.94SGTSDD354 pKa = 3.52TTPLNAKK361 pKa = 9.93VNTFLKK367 pKa = 9.61AYY369 pKa = 9.7EE370 pKa = 3.96MAAFLLSQGEE380 pKa = 4.34SAEE383 pKa = 4.43EE384 pKa = 3.65ILDD387 pKa = 3.74YY388 pKa = 10.53TYY390 pKa = 11.24HH391 pKa = 7.66DD392 pKa = 3.44WRR394 pKa = 11.84QLYY397 pKa = 8.91TMITYY402 pKa = 10.86GDD404 pKa = 3.86DD405 pKa = 3.46RR406 pKa = 11.84LVGEE410 pKa = 4.83DD411 pKa = 4.38LEE413 pKa = 5.38IPLEE417 pKa = 4.14FKK419 pKa = 10.93EE420 pKa = 4.54NIYY423 pKa = 10.8KK424 pKa = 10.05EE425 pKa = 4.02LGMWLPKK432 pKa = 10.64DD433 pKa = 3.4KK434 pKa = 10.55IKK436 pKa = 10.97RR437 pKa = 11.84QDD439 pKa = 3.55YY440 pKa = 10.51VEE442 pKa = 4.45GLSFCGAVIKK452 pKa = 10.44RR453 pKa = 11.84DD454 pKa = 3.69PCTKK458 pKa = 9.81RR459 pKa = 11.84YY460 pKa = 9.22YY461 pKa = 10.2PCFGDD466 pKa = 3.43NDD468 pKa = 3.9KK469 pKa = 11.56LIDD472 pKa = 3.72SFVWKK477 pKa = 10.4NADD480 pKa = 3.76FEE482 pKa = 4.89EE483 pKa = 4.63KK484 pKa = 10.55LLNYY488 pKa = 9.98VLISYY493 pKa = 8.67GGKK496 pKa = 9.38YY497 pKa = 7.67HH498 pKa = 7.26TSFLKK503 pKa = 8.73MAEE506 pKa = 4.18MNGVHH511 pKa = 6.25VPDD514 pKa = 4.46PQDD517 pKa = 3.03IRR519 pKa = 11.84SLIVGWW525 pKa = 3.7
Molecular weight: 61.66 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1L3KNU6|A0A1L3KNU6_9VIRU Uncharacterized protein OS=Changjiang astro-like virus OX=1922764 PE=4 SV=1
MM1 pKa = 7.39SKK3 pKa = 10.65KK4 pKa = 9.98KK5 pKa = 10.27GKK7 pKa = 10.05KK8 pKa = 8.41KK9 pKa = 10.23VVVEE13 pKa = 4.1EE14 pKa = 4.32VVVASSPTTEE24 pKa = 3.64KK25 pKa = 10.51KK26 pKa = 9.77KK27 pKa = 10.43RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84QKK32 pKa = 10.34KK33 pKa = 7.16RR34 pKa = 11.84QKK36 pKa = 9.05RR37 pKa = 11.84TAIADD42 pKa = 3.74AQAKK46 pKa = 7.16PQKK49 pKa = 10.41LPVITAYY56 pKa = 8.63STKK59 pKa = 9.6TVQISTRR66 pKa = 11.84TNKK69 pKa = 10.49NGDD72 pKa = 3.03IVMSGRR78 pKa = 11.84DD79 pKa = 3.28RR80 pKa = 11.84LEE82 pKa = 3.72VVTFPNIVTAEE93 pKa = 4.13TCVLNEE99 pKa = 3.88FLVSPKK105 pKa = 9.69YY106 pKa = 10.9FKK108 pKa = 10.66MNSRR112 pKa = 11.84LSKK115 pKa = 10.59VAANWEE121 pKa = 4.06KK122 pKa = 10.68YY123 pKa = 9.92RR124 pKa = 11.84FVKK127 pKa = 9.35FTVDD131 pKa = 3.37YY132 pKa = 10.05VPSVPSTISGQLIMSFDD149 pKa = 4.61PDD151 pKa = 3.7PEE153 pKa = 4.31GTVAGGDD160 pKa = 3.74YY161 pKa = 9.35QTVAEE166 pKa = 4.43FCMNRR171 pKa = 11.84QRR173 pKa = 11.84SVMFSINEE181 pKa = 3.98HH182 pKa = 6.03KK183 pKa = 10.27RR184 pKa = 11.84VSLNCDD190 pKa = 3.04PKK192 pKa = 11.22LGINDD197 pKa = 3.54WFVDD201 pKa = 3.98LEE203 pKa = 4.77SNGEE207 pKa = 4.04IKK209 pKa = 9.93LTAQGRR215 pKa = 11.84FDD217 pKa = 4.72LVVSTPLISGSQITQSLVVGTLFADD242 pKa = 3.59WEE244 pKa = 4.73VILKK248 pKa = 9.57QPQRR252 pKa = 11.84AQNTGAVTSRR262 pKa = 11.84SLSEE266 pKa = 3.96YY267 pKa = 10.99LNWEE271 pKa = 3.59YY272 pKa = 10.2WAPLLEE278 pKa = 4.48LSADD282 pKa = 3.41VTYY285 pKa = 10.01FNCKK289 pKa = 7.02STYY292 pKa = 10.95AVTNGFKK299 pKa = 10.71FINNSKK305 pKa = 10.2GVCEE309 pKa = 4.33TKK311 pKa = 10.75NVGYY315 pKa = 9.93VANANNGNTKK325 pKa = 10.52AGVISDD331 pKa = 3.46MMAGRR336 pKa = 11.84VYY338 pKa = 10.49VCRR341 pKa = 11.84GSFFPIGEE349 pKa = 3.83QWRR352 pKa = 11.84TYY354 pKa = 6.45TLKK357 pKa = 11.14GITVTALTRR366 pKa = 11.84NTYY369 pKa = 10.08IFFMNNFSTVDD380 pKa = 3.31NSNPLRR386 pKa = 11.84DD387 pKa = 3.7INVDD391 pKa = 3.16TDD393 pKa = 3.53SGCRR397 pKa = 11.84DD398 pKa = 2.77IVMGVRR404 pKa = 11.84FLNTQAGNMQIGYY417 pKa = 8.17YY418 pKa = 9.11TDD420 pKa = 5.07ASIQSDD426 pKa = 4.29SYY428 pKa = 9.34TRR430 pKa = 11.84QVTKK434 pKa = 10.79CGDD437 pKa = 3.48ALQGQILRR445 pKa = 11.84LLARR449 pKa = 11.84ASMWSADD456 pKa = 3.66SEE458 pKa = 4.32EE459 pKa = 4.31APRR462 pKa = 11.84LSKK465 pKa = 9.18TQIPTLEE472 pKa = 4.02DD473 pKa = 2.9RR474 pKa = 11.84LRR476 pKa = 11.84KK477 pKa = 9.34IEE479 pKa = 4.0EE480 pKa = 4.15TLDD483 pKa = 3.66VNTSRR488 pKa = 11.84YY489 pKa = 9.43EE490 pKa = 4.14GEE492 pKa = 4.23FGCC495 pKa = 6.03
MM1 pKa = 7.39SKK3 pKa = 10.65KK4 pKa = 9.98KK5 pKa = 10.27GKK7 pKa = 10.05KK8 pKa = 8.41KK9 pKa = 10.23VVVEE13 pKa = 4.1EE14 pKa = 4.32VVVASSPTTEE24 pKa = 3.64KK25 pKa = 10.51KK26 pKa = 9.77KK27 pKa = 10.43RR28 pKa = 11.84RR29 pKa = 11.84RR30 pKa = 11.84QKK32 pKa = 10.34KK33 pKa = 7.16RR34 pKa = 11.84QKK36 pKa = 9.05RR37 pKa = 11.84TAIADD42 pKa = 3.74AQAKK46 pKa = 7.16PQKK49 pKa = 10.41LPVITAYY56 pKa = 8.63STKK59 pKa = 9.6TVQISTRR66 pKa = 11.84TNKK69 pKa = 10.49NGDD72 pKa = 3.03IVMSGRR78 pKa = 11.84DD79 pKa = 3.28RR80 pKa = 11.84LEE82 pKa = 3.72VVTFPNIVTAEE93 pKa = 4.13TCVLNEE99 pKa = 3.88FLVSPKK105 pKa = 9.69YY106 pKa = 10.9FKK108 pKa = 10.66MNSRR112 pKa = 11.84LSKK115 pKa = 10.59VAANWEE121 pKa = 4.06KK122 pKa = 10.68YY123 pKa = 9.92RR124 pKa = 11.84FVKK127 pKa = 9.35FTVDD131 pKa = 3.37YY132 pKa = 10.05VPSVPSTISGQLIMSFDD149 pKa = 4.61PDD151 pKa = 3.7PEE153 pKa = 4.31GTVAGGDD160 pKa = 3.74YY161 pKa = 9.35QTVAEE166 pKa = 4.43FCMNRR171 pKa = 11.84QRR173 pKa = 11.84SVMFSINEE181 pKa = 3.98HH182 pKa = 6.03KK183 pKa = 10.27RR184 pKa = 11.84VSLNCDD190 pKa = 3.04PKK192 pKa = 11.22LGINDD197 pKa = 3.54WFVDD201 pKa = 3.98LEE203 pKa = 4.77SNGEE207 pKa = 4.04IKK209 pKa = 9.93LTAQGRR215 pKa = 11.84FDD217 pKa = 4.72LVVSTPLISGSQITQSLVVGTLFADD242 pKa = 3.59WEE244 pKa = 4.73VILKK248 pKa = 9.57QPQRR252 pKa = 11.84AQNTGAVTSRR262 pKa = 11.84SLSEE266 pKa = 3.96YY267 pKa = 10.99LNWEE271 pKa = 3.59YY272 pKa = 10.2WAPLLEE278 pKa = 4.48LSADD282 pKa = 3.41VTYY285 pKa = 10.01FNCKK289 pKa = 7.02STYY292 pKa = 10.95AVTNGFKK299 pKa = 10.71FINNSKK305 pKa = 10.2GVCEE309 pKa = 4.33TKK311 pKa = 10.75NVGYY315 pKa = 9.93VANANNGNTKK325 pKa = 10.52AGVISDD331 pKa = 3.46MMAGRR336 pKa = 11.84VYY338 pKa = 10.49VCRR341 pKa = 11.84GSFFPIGEE349 pKa = 3.83QWRR352 pKa = 11.84TYY354 pKa = 6.45TLKK357 pKa = 11.14GITVTALTRR366 pKa = 11.84NTYY369 pKa = 10.08IFFMNNFSTVDD380 pKa = 3.31NSNPLRR386 pKa = 11.84DD387 pKa = 3.7INVDD391 pKa = 3.16TDD393 pKa = 3.53SGCRR397 pKa = 11.84DD398 pKa = 2.77IVMGVRR404 pKa = 11.84FLNTQAGNMQIGYY417 pKa = 8.17YY418 pKa = 9.11TDD420 pKa = 5.07ASIQSDD426 pKa = 4.29SYY428 pKa = 9.34TRR430 pKa = 11.84QVTKK434 pKa = 10.79CGDD437 pKa = 3.48ALQGQILRR445 pKa = 11.84LLARR449 pKa = 11.84ASMWSADD456 pKa = 3.66SEE458 pKa = 4.32EE459 pKa = 4.31APRR462 pKa = 11.84LSKK465 pKa = 9.18TQIPTLEE472 pKa = 4.02DD473 pKa = 2.9RR474 pKa = 11.84LRR476 pKa = 11.84KK477 pKa = 9.34IEE479 pKa = 4.0EE480 pKa = 4.15TLDD483 pKa = 3.66VNTSRR488 pKa = 11.84YY489 pKa = 9.43EE490 pKa = 4.14GEE492 pKa = 4.23FGCC495 pKa = 6.03
Molecular weight: 55.47 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2102 |
302 |
780 |
525.5 |
60.03 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.328 ± 0.34 | 2.521 ± 0.701 |
5.947 ± 0.326 | 5.804 ± 0.791 |
3.092 ± 0.484 | 6.518 ± 0.618 |
2.426 ± 0.569 | 5.852 ± 0.277 |
5.899 ± 0.926 | 8.23 ± 0.554 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.997 ± 0.483 | 4.757 ± 0.617 |
4.044 ± 0.305 | 3.425 ± 0.391 |
5.804 ± 0.738 | 6.137 ± 0.497 |
6.185 ± 0.754 | 7.945 ± 0.741 |
2.569 ± 0.415 | 4.52 ± 0.515 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
