
Faeces associated gemycircularvirus 10
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemygorvirus; Gemygorvirus stara1; Starling associated gemygorvirus 1
Average proteome isoelectric point is 7.14
Get precalculated fractions of proteins
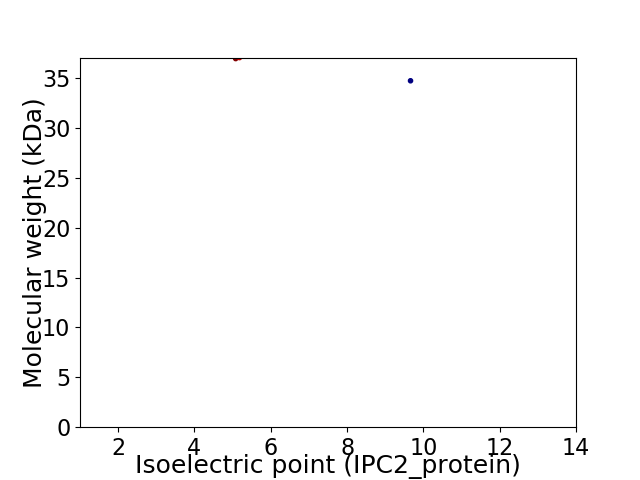
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|T1YRV6|T1YRV6_9VIRU Capsid protein OS=Faeces associated gemycircularvirus 10 OX=1391025 PE=4 SV=1
MM1 pKa = 7.5SNSTLLSVTLCPHH14 pKa = 6.03YY15 pKa = 10.8LCPCDD20 pKa = 5.08GLDD23 pKa = 3.19YY24 pKa = 10.83WRR26 pKa = 11.84VLDD29 pKa = 4.57HH30 pKa = 6.78LSEE33 pKa = 4.43VPAEE37 pKa = 4.16CIIGRR42 pKa = 11.84EE43 pKa = 3.97AHH45 pKa = 6.81ADD47 pKa = 3.53GGTHH51 pKa = 6.7LHH53 pKa = 6.66AFCDD57 pKa = 4.23FGEE60 pKa = 4.35RR61 pKa = 11.84FYY63 pKa = 10.87TADD66 pKa = 3.36PRR68 pKa = 11.84RR69 pKa = 11.84FDD71 pKa = 3.24VDD73 pKa = 4.03GYY75 pKa = 10.65HH76 pKa = 7.43PNVQPFGRR84 pKa = 11.84TPYY87 pKa = 10.29KK88 pKa = 10.43GWDD91 pKa = 3.43YY92 pKa = 11.11AIKK95 pKa = 10.84DD96 pKa = 3.55GDD98 pKa = 4.3VVCGGLEE105 pKa = 3.97RR106 pKa = 11.84PGTGPGTDD114 pKa = 3.02VSGAGSQWDD123 pKa = 4.01RR124 pKa = 11.84IVGAEE129 pKa = 4.23TPDD132 pKa = 3.51EE133 pKa = 4.15FWRR136 pKa = 11.84LIRR139 pKa = 11.84EE140 pKa = 4.38LAPRR144 pKa = 11.84TLLTNFNSVRR154 pKa = 11.84AYY156 pKa = 10.65ADD158 pKa = 2.69WHH160 pKa = 5.55YY161 pKa = 10.75RR162 pKa = 11.84PVVAPYY168 pKa = 9.03EE169 pKa = 4.33SPPEE173 pKa = 3.9IEE175 pKa = 4.44FDD177 pKa = 3.48LSGVEE182 pKa = 4.74EE183 pKa = 4.05LRR185 pKa = 11.84EE186 pKa = 4.09WVCNNLSRR194 pKa = 11.84TRR196 pKa = 11.84RR197 pKa = 11.84PKK199 pKa = 10.61SLVLYY204 pKa = 10.61GEE206 pKa = 4.24SRR208 pKa = 11.84LGKK211 pKa = 7.36TLWARR216 pKa = 11.84SLGRR220 pKa = 11.84HH221 pKa = 5.63VYY223 pKa = 10.33NCLQFNVDD231 pKa = 3.92DD232 pKa = 3.77MRR234 pKa = 11.84GDD236 pKa = 3.52VEE238 pKa = 4.03EE239 pKa = 4.36ALYY242 pKa = 10.91AVFDD246 pKa = 4.22DD247 pKa = 3.87MQGGFKK253 pKa = 10.6YY254 pKa = 10.49FPSYY258 pKa = 10.47KK259 pKa = 9.96GWLGAQSKK267 pKa = 7.6FTVTDD272 pKa = 3.76KK273 pKa = 11.37YY274 pKa = 10.65RR275 pKa = 11.84GKK277 pKa = 9.31TSVIWGRR284 pKa = 11.84PTIWLCNEE292 pKa = 3.98SPRR295 pKa = 11.84DD296 pKa = 3.75FTDD299 pKa = 4.49VDD301 pKa = 4.2MTWLEE306 pKa = 4.1ANCVFVEE313 pKa = 4.07VTNPIFRR320 pKa = 11.84ANIEE324 pKa = 3.95
MM1 pKa = 7.5SNSTLLSVTLCPHH14 pKa = 6.03YY15 pKa = 10.8LCPCDD20 pKa = 5.08GLDD23 pKa = 3.19YY24 pKa = 10.83WRR26 pKa = 11.84VLDD29 pKa = 4.57HH30 pKa = 6.78LSEE33 pKa = 4.43VPAEE37 pKa = 4.16CIIGRR42 pKa = 11.84EE43 pKa = 3.97AHH45 pKa = 6.81ADD47 pKa = 3.53GGTHH51 pKa = 6.7LHH53 pKa = 6.66AFCDD57 pKa = 4.23FGEE60 pKa = 4.35RR61 pKa = 11.84FYY63 pKa = 10.87TADD66 pKa = 3.36PRR68 pKa = 11.84RR69 pKa = 11.84FDD71 pKa = 3.24VDD73 pKa = 4.03GYY75 pKa = 10.65HH76 pKa = 7.43PNVQPFGRR84 pKa = 11.84TPYY87 pKa = 10.29KK88 pKa = 10.43GWDD91 pKa = 3.43YY92 pKa = 11.11AIKK95 pKa = 10.84DD96 pKa = 3.55GDD98 pKa = 4.3VVCGGLEE105 pKa = 3.97RR106 pKa = 11.84PGTGPGTDD114 pKa = 3.02VSGAGSQWDD123 pKa = 4.01RR124 pKa = 11.84IVGAEE129 pKa = 4.23TPDD132 pKa = 3.51EE133 pKa = 4.15FWRR136 pKa = 11.84LIRR139 pKa = 11.84EE140 pKa = 4.38LAPRR144 pKa = 11.84TLLTNFNSVRR154 pKa = 11.84AYY156 pKa = 10.65ADD158 pKa = 2.69WHH160 pKa = 5.55YY161 pKa = 10.75RR162 pKa = 11.84PVVAPYY168 pKa = 9.03EE169 pKa = 4.33SPPEE173 pKa = 3.9IEE175 pKa = 4.44FDD177 pKa = 3.48LSGVEE182 pKa = 4.74EE183 pKa = 4.05LRR185 pKa = 11.84EE186 pKa = 4.09WVCNNLSRR194 pKa = 11.84TRR196 pKa = 11.84RR197 pKa = 11.84PKK199 pKa = 10.61SLVLYY204 pKa = 10.61GEE206 pKa = 4.24SRR208 pKa = 11.84LGKK211 pKa = 7.36TLWARR216 pKa = 11.84SLGRR220 pKa = 11.84HH221 pKa = 5.63VYY223 pKa = 10.33NCLQFNVDD231 pKa = 3.92DD232 pKa = 3.77MRR234 pKa = 11.84GDD236 pKa = 3.52VEE238 pKa = 4.03EE239 pKa = 4.36ALYY242 pKa = 10.91AVFDD246 pKa = 4.22DD247 pKa = 3.87MQGGFKK253 pKa = 10.6YY254 pKa = 10.49FPSYY258 pKa = 10.47KK259 pKa = 9.96GWLGAQSKK267 pKa = 7.6FTVTDD272 pKa = 3.76KK273 pKa = 11.37YY274 pKa = 10.65RR275 pKa = 11.84GKK277 pKa = 9.31TSVIWGRR284 pKa = 11.84PTIWLCNEE292 pKa = 3.98SPRR295 pKa = 11.84DD296 pKa = 3.75FTDD299 pKa = 4.49VDD301 pKa = 4.2MTWLEE306 pKa = 4.1ANCVFVEE313 pKa = 4.07VTNPIFRR320 pKa = 11.84ANIEE324 pKa = 3.95
Molecular weight: 36.95 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|T1YRV6|T1YRV6_9VIRU Capsid protein OS=Faeces associated gemycircularvirus 10 OX=1391025 PE=4 SV=1
MM1 pKa = 7.54PGYY4 pKa = 9.94RR5 pKa = 11.84SKK7 pKa = 11.0RR8 pKa = 11.84KK9 pKa = 9.26YY10 pKa = 10.18RR11 pKa = 11.84RR12 pKa = 11.84TSRR15 pKa = 11.84KK16 pKa = 8.74GGRR19 pKa = 11.84SLRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84SGYY27 pKa = 9.84RR28 pKa = 11.84KK29 pKa = 8.82SSKK32 pKa = 9.72GRR34 pKa = 11.84RR35 pKa = 11.84FLRR38 pKa = 11.84GRR40 pKa = 11.84RR41 pKa = 11.84GMSRR45 pKa = 11.84KK46 pKa = 9.7ALLNVTSRR54 pKa = 11.84KK55 pKa = 9.78KK56 pKa = 10.19RR57 pKa = 11.84DD58 pKa = 3.21TMLSYY63 pKa = 11.18TNVSTSSQIGSANYY77 pKa = 9.47VVGPAVLAGGAQRR90 pKa = 11.84FFLWCATARR99 pKa = 11.84GISTYY104 pKa = 11.36NNFDD108 pKa = 3.41ASLIDD113 pKa = 3.6QSSRR117 pKa = 11.84TATNCYY123 pKa = 9.02MRR125 pKa = 11.84GLKK128 pKa = 9.77EE129 pKa = 4.37RR130 pKa = 11.84IRR132 pKa = 11.84ISTDD136 pKa = 3.54DD137 pKa = 3.51GMPWLWRR144 pKa = 11.84RR145 pKa = 11.84ICFTSRR151 pKa = 11.84NLNLRR156 pKa = 11.84GYY158 pKa = 7.87QTGAGTSYY166 pKa = 11.29SPAFLTSAGFMRR178 pKa = 11.84GVNQPYY184 pKa = 9.6NDD186 pKa = 3.74SYY188 pKa = 11.8INDD191 pKa = 4.57LLWKK195 pKa = 8.89GTLGKK200 pKa = 10.11DD201 pKa = 2.85WNDD204 pKa = 3.0FMTATVDD211 pKa = 3.29NRR213 pKa = 11.84RR214 pKa = 11.84VDD216 pKa = 3.51VKK218 pKa = 10.96YY219 pKa = 11.27DD220 pKa = 3.2KK221 pKa = 8.68TTTIAAGNEE230 pKa = 4.02EE231 pKa = 4.21GCIRR235 pKa = 11.84EE236 pKa = 4.09FNRR239 pKa = 11.84WHH241 pKa = 6.97PMNKK245 pKa = 9.63SLVYY249 pKa = 10.49DD250 pKa = 3.94DD251 pKa = 5.57EE252 pKa = 4.5EE253 pKa = 6.46AGDD256 pKa = 3.79EE257 pKa = 4.15AAGSVFSVTDD267 pKa = 3.22KK268 pKa = 11.26RR269 pKa = 11.84GMGDD273 pKa = 3.94YY274 pKa = 11.04YY275 pKa = 11.65VMDD278 pKa = 3.57IFDD281 pKa = 4.56PRR283 pKa = 11.84SGSTSASHH291 pKa = 7.44LSFRR295 pKa = 11.84PDD297 pKa = 2.7ATLYY301 pKa = 7.81WHH303 pKa = 6.64EE304 pKa = 4.21RR305 pKa = 3.47
MM1 pKa = 7.54PGYY4 pKa = 9.94RR5 pKa = 11.84SKK7 pKa = 11.0RR8 pKa = 11.84KK9 pKa = 9.26YY10 pKa = 10.18RR11 pKa = 11.84RR12 pKa = 11.84TSRR15 pKa = 11.84KK16 pKa = 8.74GGRR19 pKa = 11.84SLRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84SGYY27 pKa = 9.84RR28 pKa = 11.84KK29 pKa = 8.82SSKK32 pKa = 9.72GRR34 pKa = 11.84RR35 pKa = 11.84FLRR38 pKa = 11.84GRR40 pKa = 11.84RR41 pKa = 11.84GMSRR45 pKa = 11.84KK46 pKa = 9.7ALLNVTSRR54 pKa = 11.84KK55 pKa = 9.78KK56 pKa = 10.19RR57 pKa = 11.84DD58 pKa = 3.21TMLSYY63 pKa = 11.18TNVSTSSQIGSANYY77 pKa = 9.47VVGPAVLAGGAQRR90 pKa = 11.84FFLWCATARR99 pKa = 11.84GISTYY104 pKa = 11.36NNFDD108 pKa = 3.41ASLIDD113 pKa = 3.6QSSRR117 pKa = 11.84TATNCYY123 pKa = 9.02MRR125 pKa = 11.84GLKK128 pKa = 9.77EE129 pKa = 4.37RR130 pKa = 11.84IRR132 pKa = 11.84ISTDD136 pKa = 3.54DD137 pKa = 3.51GMPWLWRR144 pKa = 11.84RR145 pKa = 11.84ICFTSRR151 pKa = 11.84NLNLRR156 pKa = 11.84GYY158 pKa = 7.87QTGAGTSYY166 pKa = 11.29SPAFLTSAGFMRR178 pKa = 11.84GVNQPYY184 pKa = 9.6NDD186 pKa = 3.74SYY188 pKa = 11.8INDD191 pKa = 4.57LLWKK195 pKa = 8.89GTLGKK200 pKa = 10.11DD201 pKa = 2.85WNDD204 pKa = 3.0FMTATVDD211 pKa = 3.29NRR213 pKa = 11.84RR214 pKa = 11.84VDD216 pKa = 3.51VKK218 pKa = 10.96YY219 pKa = 11.27DD220 pKa = 3.2KK221 pKa = 8.68TTTIAAGNEE230 pKa = 4.02EE231 pKa = 4.21GCIRR235 pKa = 11.84EE236 pKa = 4.09FNRR239 pKa = 11.84WHH241 pKa = 6.97PMNKK245 pKa = 9.63SLVYY249 pKa = 10.49DD250 pKa = 3.94DD251 pKa = 5.57EE252 pKa = 4.5EE253 pKa = 6.46AGDD256 pKa = 3.79EE257 pKa = 4.15AAGSVFSVTDD267 pKa = 3.22KK268 pKa = 11.26RR269 pKa = 11.84GMGDD273 pKa = 3.94YY274 pKa = 11.04YY275 pKa = 11.65VMDD278 pKa = 3.57IFDD281 pKa = 4.56PRR283 pKa = 11.84SGSTSASHH291 pKa = 7.44LSFRR295 pKa = 11.84PDD297 pKa = 2.7ATLYY301 pKa = 7.81WHH303 pKa = 6.64EE304 pKa = 4.21RR305 pKa = 3.47
Molecular weight: 34.73 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
629 |
305 |
324 |
314.5 |
35.84 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.041 ± 0.358 | 2.226 ± 0.635 |
7.154 ± 0.414 | 4.61 ± 1.38 |
4.452 ± 0.359 | 8.903 ± 0.193 |
1.749 ± 0.531 | 3.18 ± 0.069 |
3.816 ± 0.765 | 7.154 ± 0.642 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.226 ± 0.731 | 4.452 ± 0.551 |
4.452 ± 1.269 | 1.59 ± 0.034 |
9.857 ± 1.579 | 7.631 ± 1.758 |
6.677 ± 0.6 | 6.041 ± 1.235 |
2.862 ± 0.393 | 4.928 ± 0.22 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
