
Acetohalobium arabaticum (strain ATCC 49924 / DSM 5501 / Z-7288)
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Clostridia; Halanaerobiales; Halobacteroidaceae; Acetohalobium; Acetohalobium arabaticum
Average proteome isoelectric point is 5.69
Get precalculated fractions of proteins
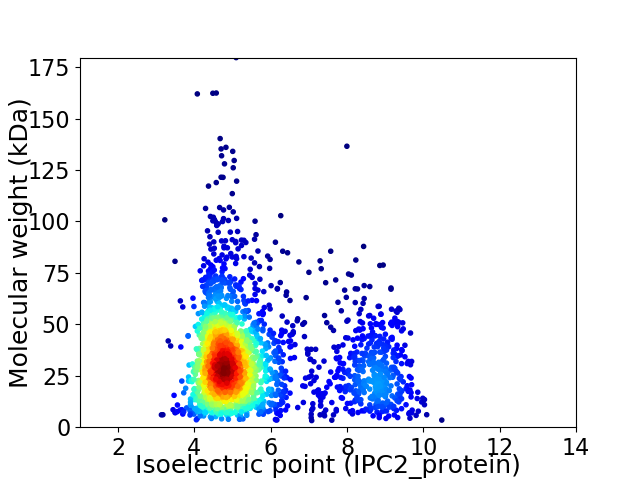
Virtual 2D-PAGE plot for 2276 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|D9QSU2|D9QSU2_ACEAZ Ribonuclease J OS=Acetohalobium arabaticum (strain ATCC 49924 / DSM 5501 / Z-7288) OX=574087 GN=rnj PE=3 SV=1
MM1 pKa = 7.66SKK3 pKa = 10.78FNLQEE8 pKa = 4.17KK9 pKa = 10.23NNSNVNGNDD18 pKa = 3.32FAVEE22 pKa = 4.35VPFCCVQVVPGNLEE36 pKa = 3.98VVEE39 pKa = 4.76DD40 pKa = 3.86RR41 pKa = 11.84LEE43 pKa = 5.35AEE45 pKa = 4.29DD46 pKa = 5.1LKK48 pKa = 9.59MTFTPKK54 pKa = 10.5LEE56 pKa = 4.2CCIKK60 pKa = 10.85EE61 pKa = 4.03EE62 pKa = 4.43DD63 pKa = 4.14LSCIADD69 pKa = 3.7NVSCSIDD76 pKa = 3.72GEE78 pKa = 4.36VLRR81 pKa = 11.84LVGCIEE87 pKa = 4.11YY88 pKa = 10.41AISTINGTPDD98 pKa = 3.31PVRR101 pKa = 11.84GDD103 pKa = 4.02FSTEE107 pKa = 3.42APQQNSADD115 pKa = 3.9VSCKK119 pKa = 9.61STVCVNEE126 pKa = 4.81IICCGQDD133 pKa = 3.79LEE135 pKa = 5.54CPDD138 pKa = 5.51DD139 pKa = 5.02LDD141 pKa = 5.04LNEE144 pKa = 4.0ITVNFEE150 pKa = 3.63DD151 pKa = 5.48AKK153 pKa = 11.56VKK155 pKa = 10.52DD156 pKa = 3.86CPKK159 pKa = 10.18WADD162 pKa = 3.69EE163 pKa = 4.12PDD165 pKa = 3.19KK166 pKa = 11.28KK167 pKa = 10.51IIKK170 pKa = 8.65FTGKK174 pKa = 9.7FRR176 pKa = 11.84LPTLPTCPDD185 pKa = 3.16EE186 pKa = 4.61NGNGNGDD193 pKa = 3.59GNGNGNGNGNDD204 pKa = 3.53NGDD207 pKa = 3.71NGDD210 pKa = 4.25EE211 pKa = 4.33EE212 pKa = 5.38VCGCQVTGDD221 pKa = 4.43LNTGGNPDD229 pKa = 3.46EE230 pKa = 5.27RR231 pKa = 11.84VDD233 pKa = 5.87LNLQVCTGCTPEE245 pKa = 4.24NSLVSYY251 pKa = 11.39NNTLEE256 pKa = 5.13DD257 pKa = 4.07NDD259 pKa = 3.85IQLDD263 pKa = 4.11LSPDD267 pKa = 3.53DD268 pKa = 4.93EE269 pKa = 4.51EE270 pKa = 5.95PEE272 pKa = 5.15GIDD275 pKa = 4.74LVTCEE280 pKa = 4.86EE281 pKa = 3.87IAEE284 pKa = 4.25IEE286 pKa = 4.28GGLRR290 pKa = 11.84ATIEE294 pKa = 4.23GQASVTVGEE303 pKa = 4.94DD304 pKa = 2.83SFAACDD310 pKa = 3.71FTIVVTDD317 pKa = 4.1TPPPTSDD324 pKa = 3.05TIEE327 pKa = 4.07EE328 pKa = 4.17VEE330 pKa = 5.18IICDD334 pKa = 3.73DD335 pKa = 3.84QSVHH339 pKa = 5.47TVTEE343 pKa = 4.82PIEE346 pKa = 4.13GAQAINIQDD355 pKa = 3.66ICPLPPQQQ363 pKa = 4.62
MM1 pKa = 7.66SKK3 pKa = 10.78FNLQEE8 pKa = 4.17KK9 pKa = 10.23NNSNVNGNDD18 pKa = 3.32FAVEE22 pKa = 4.35VPFCCVQVVPGNLEE36 pKa = 3.98VVEE39 pKa = 4.76DD40 pKa = 3.86RR41 pKa = 11.84LEE43 pKa = 5.35AEE45 pKa = 4.29DD46 pKa = 5.1LKK48 pKa = 9.59MTFTPKK54 pKa = 10.5LEE56 pKa = 4.2CCIKK60 pKa = 10.85EE61 pKa = 4.03EE62 pKa = 4.43DD63 pKa = 4.14LSCIADD69 pKa = 3.7NVSCSIDD76 pKa = 3.72GEE78 pKa = 4.36VLRR81 pKa = 11.84LVGCIEE87 pKa = 4.11YY88 pKa = 10.41AISTINGTPDD98 pKa = 3.31PVRR101 pKa = 11.84GDD103 pKa = 4.02FSTEE107 pKa = 3.42APQQNSADD115 pKa = 3.9VSCKK119 pKa = 9.61STVCVNEE126 pKa = 4.81IICCGQDD133 pKa = 3.79LEE135 pKa = 5.54CPDD138 pKa = 5.51DD139 pKa = 5.02LDD141 pKa = 5.04LNEE144 pKa = 4.0ITVNFEE150 pKa = 3.63DD151 pKa = 5.48AKK153 pKa = 11.56VKK155 pKa = 10.52DD156 pKa = 3.86CPKK159 pKa = 10.18WADD162 pKa = 3.69EE163 pKa = 4.12PDD165 pKa = 3.19KK166 pKa = 11.28KK167 pKa = 10.51IIKK170 pKa = 8.65FTGKK174 pKa = 9.7FRR176 pKa = 11.84LPTLPTCPDD185 pKa = 3.16EE186 pKa = 4.61NGNGNGDD193 pKa = 3.59GNGNGNGNGNDD204 pKa = 3.53NGDD207 pKa = 3.71NGDD210 pKa = 4.25EE211 pKa = 4.33EE212 pKa = 5.38VCGCQVTGDD221 pKa = 4.43LNTGGNPDD229 pKa = 3.46EE230 pKa = 5.27RR231 pKa = 11.84VDD233 pKa = 5.87LNLQVCTGCTPEE245 pKa = 4.24NSLVSYY251 pKa = 11.39NNTLEE256 pKa = 5.13DD257 pKa = 4.07NDD259 pKa = 3.85IQLDD263 pKa = 4.11LSPDD267 pKa = 3.53DD268 pKa = 4.93EE269 pKa = 4.51EE270 pKa = 5.95PEE272 pKa = 5.15GIDD275 pKa = 4.74LVTCEE280 pKa = 4.86EE281 pKa = 3.87IAEE284 pKa = 4.25IEE286 pKa = 4.28GGLRR290 pKa = 11.84ATIEE294 pKa = 4.23GQASVTVGEE303 pKa = 4.94DD304 pKa = 2.83SFAACDD310 pKa = 3.71FTIVVTDD317 pKa = 4.1TPPPTSDD324 pKa = 3.05TIEE327 pKa = 4.07EE328 pKa = 4.17VEE330 pKa = 5.18IICDD334 pKa = 3.73DD335 pKa = 3.84QSVHH339 pKa = 5.47TVTEE343 pKa = 4.82PIEE346 pKa = 4.13GAQAINIQDD355 pKa = 3.66ICPLPPQQQ363 pKa = 4.62
Molecular weight: 39.01 kDa
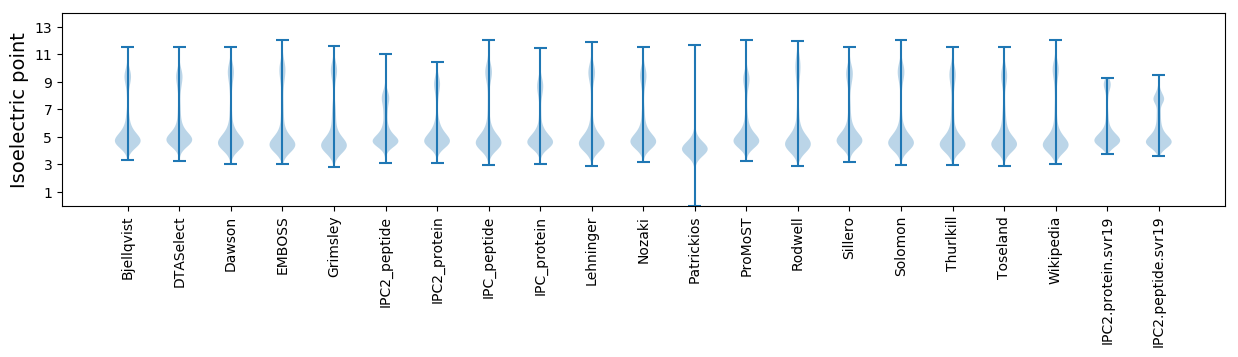
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|D9QPI9|D9QPI9_ACEAZ Dihydroorotase OS=Acetohalobium arabaticum (strain ATCC 49924 / DSM 5501 / Z-7288) OX=574087 GN=pyrC PE=3 SV=1
MM1 pKa = 7.12YY2 pKa = 10.92ALIRR6 pKa = 11.84LIDD9 pKa = 4.14LSFTIYY15 pKa = 9.3TWILIARR22 pKa = 11.84VISSWVSPPMHH33 pKa = 6.64NSNVRR38 pKa = 11.84KK39 pKa = 9.36IMKK42 pKa = 9.94FIYY45 pKa = 10.05EE46 pKa = 3.9VTEE49 pKa = 3.93PVLAPIRR56 pKa = 11.84RR57 pKa = 11.84MLPTGNIGIDD67 pKa = 4.17LSPLIAFIAINIIHH81 pKa = 6.61NSLLRR86 pKa = 11.84ILRR89 pKa = 11.84RR90 pKa = 11.84LLLYY94 pKa = 10.65
MM1 pKa = 7.12YY2 pKa = 10.92ALIRR6 pKa = 11.84LIDD9 pKa = 4.14LSFTIYY15 pKa = 9.3TWILIARR22 pKa = 11.84VISSWVSPPMHH33 pKa = 6.64NSNVRR38 pKa = 11.84KK39 pKa = 9.36IMKK42 pKa = 9.94FIYY45 pKa = 10.05EE46 pKa = 3.9VTEE49 pKa = 3.93PVLAPIRR56 pKa = 11.84RR57 pKa = 11.84MLPTGNIGIDD67 pKa = 4.17LSPLIAFIAINIIHH81 pKa = 6.61NSLLRR86 pKa = 11.84ILRR89 pKa = 11.84RR90 pKa = 11.84LLLYY94 pKa = 10.65
Molecular weight: 10.92 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
703237 |
30 |
1535 |
309.0 |
34.58 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.788 ± 0.052 | 0.854 ± 0.023 |
6.15 ± 0.048 | 9.025 ± 0.064 |
3.67 ± 0.033 | 6.976 ± 0.046 |
1.456 ± 0.02 | 8.313 ± 0.049 |
7.104 ± 0.057 | 10.106 ± 0.063 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.249 ± 0.022 | 4.686 ± 0.036 |
3.297 ± 0.028 | 3.63 ± 0.033 |
4.002 ± 0.033 | 5.5 ± 0.036 |
5.008 ± 0.032 | 7.15 ± 0.045 |
0.717 ± 0.016 | 3.319 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
