
Human papillomavirus type 131
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Gammapapillomavirus; Gammapapillomavirus 14
Average proteome isoelectric point is 6.16
Get precalculated fractions of proteins
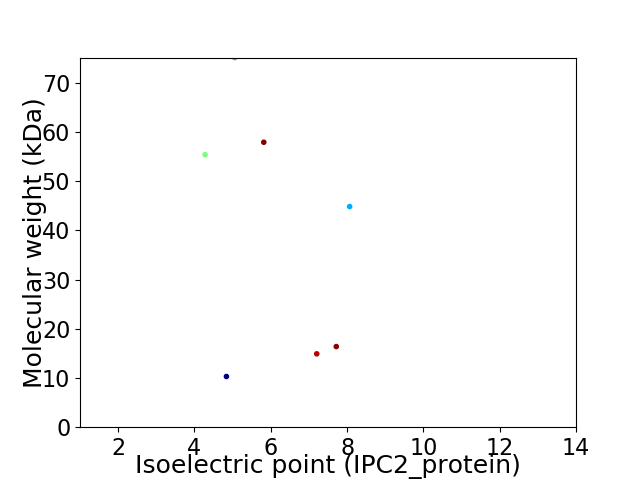
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|E7BQ92|E7BQ92_9PAPI Major capsid protein L1 OS=Human papillomavirus type 131 OX=909330 GN=L1 PE=3 SV=1
MM1 pKa = 7.4SKK3 pKa = 9.74IRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.7RR8 pKa = 11.84ASPSDD13 pKa = 3.79LYY15 pKa = 10.09NTCLQGGDD23 pKa = 4.08CVPDD27 pKa = 3.43VKK29 pKa = 11.27NKK31 pKa = 10.33FEE33 pKa = 4.95NNTIADD39 pKa = 3.96WLLKK43 pKa = 10.07IFGSIVYY50 pKa = 9.49FGNLGIGTGRR60 pKa = 11.84GTGGLFGYY68 pKa = 10.36RR69 pKa = 11.84PFGSANAGKK78 pKa = 9.02PIQEE82 pKa = 4.27LPITRR87 pKa = 11.84PNVVIEE93 pKa = 4.73PIGPTPIIPVEE104 pKa = 4.24PGASSIVPLVEE115 pKa = 3.79GTPDD119 pKa = 2.9ISFISPDD126 pKa = 3.35AGPGVGGEE134 pKa = 4.27DD135 pKa = 3.02IEE137 pKa = 5.45LFTISDD143 pKa = 3.81PTRR146 pKa = 11.84DD147 pKa = 3.12IGGVGGGPTVISTEE161 pKa = 3.97EE162 pKa = 4.16SEE164 pKa = 4.44TTIIDD169 pKa = 4.16AFPSPAGPKK178 pKa = 9.37QVFYY182 pKa = 11.15DD183 pKa = 3.31AATNITLEE191 pKa = 4.2TQINPFINADD201 pKa = 3.35INNTNIFVDD210 pKa = 3.61TTIAGEE216 pKa = 4.43TVGDD220 pKa = 3.81PSFEE224 pKa = 4.66NIPLEE229 pKa = 4.23RR230 pKa = 11.84LDD232 pKa = 3.66LQEE235 pKa = 5.25FEE237 pKa = 4.78IEE239 pKa = 4.19EE240 pKa = 4.79PPTEE244 pKa = 4.33STPVTAFGRR253 pKa = 11.84AFNRR257 pKa = 11.84ARR259 pKa = 11.84DD260 pKa = 3.65LYY262 pKa = 11.26SRR264 pKa = 11.84FIEE267 pKa = 3.83QVPITEE273 pKa = 4.67PEE275 pKa = 4.19FLVQPSRR282 pKa = 11.84LVQFEE287 pKa = 4.47SEE289 pKa = 4.47NPAFDD294 pKa = 4.94PDD296 pKa = 3.25VSLAFEE302 pKa = 4.59HH303 pKa = 7.5DD304 pKa = 3.83VAEE307 pKa = 4.37LQAAPSSEE315 pKa = 3.76FADD318 pKa = 4.22IIRR321 pKa = 11.84LSRR324 pKa = 11.84PRR326 pKa = 11.84LTATVEE332 pKa = 3.91GTVRR336 pKa = 11.84VSRR339 pKa = 11.84LGTRR343 pKa = 11.84AALTTRR349 pKa = 11.84SGLTVGPQVHH359 pKa = 6.99FYY361 pKa = 10.26MDD363 pKa = 4.19LSEE366 pKa = 4.61IAAAEE371 pKa = 4.33NIEE374 pKa = 4.09MLPIAEE380 pKa = 4.49YY381 pKa = 9.68SHH383 pKa = 6.89EE384 pKa = 4.29STVVDD389 pKa = 4.41DD390 pKa = 5.55LLAHH394 pKa = 5.68TVIDD398 pKa = 4.5DD399 pKa = 3.61VANAANINYY408 pKa = 8.71TEE410 pKa = 4.9ADD412 pKa = 3.83LEE414 pKa = 4.47DD415 pKa = 4.35LLSEE419 pKa = 4.27QFNNSHH425 pKa = 6.29IVVTSTTEE433 pKa = 3.57EE434 pKa = 4.81GEE436 pKa = 4.32TLNIPTLPPNAVLKK450 pKa = 10.73VFVPDD455 pKa = 3.31VGDD458 pKa = 3.9GLFVAYY464 pKa = 9.93GVDD467 pKa = 3.31TASNTNIVLPNNLPVEE483 pKa = 4.17PSLIITDD490 pKa = 3.42SHH492 pKa = 8.13DD493 pKa = 4.04FEE495 pKa = 5.07LHH497 pKa = 5.44PALWPRR503 pKa = 11.84KK504 pKa = 8.67RR505 pKa = 11.84RR506 pKa = 11.84RR507 pKa = 11.84LDD509 pKa = 3.23LFF511 pKa = 4.04
MM1 pKa = 7.4SKK3 pKa = 9.74IRR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 9.7RR8 pKa = 11.84ASPSDD13 pKa = 3.79LYY15 pKa = 10.09NTCLQGGDD23 pKa = 4.08CVPDD27 pKa = 3.43VKK29 pKa = 11.27NKK31 pKa = 10.33FEE33 pKa = 4.95NNTIADD39 pKa = 3.96WLLKK43 pKa = 10.07IFGSIVYY50 pKa = 9.49FGNLGIGTGRR60 pKa = 11.84GTGGLFGYY68 pKa = 10.36RR69 pKa = 11.84PFGSANAGKK78 pKa = 9.02PIQEE82 pKa = 4.27LPITRR87 pKa = 11.84PNVVIEE93 pKa = 4.73PIGPTPIIPVEE104 pKa = 4.24PGASSIVPLVEE115 pKa = 3.79GTPDD119 pKa = 2.9ISFISPDD126 pKa = 3.35AGPGVGGEE134 pKa = 4.27DD135 pKa = 3.02IEE137 pKa = 5.45LFTISDD143 pKa = 3.81PTRR146 pKa = 11.84DD147 pKa = 3.12IGGVGGGPTVISTEE161 pKa = 3.97EE162 pKa = 4.16SEE164 pKa = 4.44TTIIDD169 pKa = 4.16AFPSPAGPKK178 pKa = 9.37QVFYY182 pKa = 11.15DD183 pKa = 3.31AATNITLEE191 pKa = 4.2TQINPFINADD201 pKa = 3.35INNTNIFVDD210 pKa = 3.61TTIAGEE216 pKa = 4.43TVGDD220 pKa = 3.81PSFEE224 pKa = 4.66NIPLEE229 pKa = 4.23RR230 pKa = 11.84LDD232 pKa = 3.66LQEE235 pKa = 5.25FEE237 pKa = 4.78IEE239 pKa = 4.19EE240 pKa = 4.79PPTEE244 pKa = 4.33STPVTAFGRR253 pKa = 11.84AFNRR257 pKa = 11.84ARR259 pKa = 11.84DD260 pKa = 3.65LYY262 pKa = 11.26SRR264 pKa = 11.84FIEE267 pKa = 3.83QVPITEE273 pKa = 4.67PEE275 pKa = 4.19FLVQPSRR282 pKa = 11.84LVQFEE287 pKa = 4.47SEE289 pKa = 4.47NPAFDD294 pKa = 4.94PDD296 pKa = 3.25VSLAFEE302 pKa = 4.59HH303 pKa = 7.5DD304 pKa = 3.83VAEE307 pKa = 4.37LQAAPSSEE315 pKa = 3.76FADD318 pKa = 4.22IIRR321 pKa = 11.84LSRR324 pKa = 11.84PRR326 pKa = 11.84LTATVEE332 pKa = 3.91GTVRR336 pKa = 11.84VSRR339 pKa = 11.84LGTRR343 pKa = 11.84AALTTRR349 pKa = 11.84SGLTVGPQVHH359 pKa = 6.99FYY361 pKa = 10.26MDD363 pKa = 4.19LSEE366 pKa = 4.61IAAAEE371 pKa = 4.33NIEE374 pKa = 4.09MLPIAEE380 pKa = 4.49YY381 pKa = 9.68SHH383 pKa = 6.89EE384 pKa = 4.29STVVDD389 pKa = 4.41DD390 pKa = 5.55LLAHH394 pKa = 5.68TVIDD398 pKa = 4.5DD399 pKa = 3.61VANAANINYY408 pKa = 8.71TEE410 pKa = 4.9ADD412 pKa = 3.83LEE414 pKa = 4.47DD415 pKa = 4.35LLSEE419 pKa = 4.27QFNNSHH425 pKa = 6.29IVVTSTTEE433 pKa = 3.57EE434 pKa = 4.81GEE436 pKa = 4.32TLNIPTLPPNAVLKK450 pKa = 10.73VFVPDD455 pKa = 3.31VGDD458 pKa = 3.9GLFVAYY464 pKa = 9.93GVDD467 pKa = 3.31TASNTNIVLPNNLPVEE483 pKa = 4.17PSLIITDD490 pKa = 3.42SHH492 pKa = 8.13DD493 pKa = 4.04FEE495 pKa = 5.07LHH497 pKa = 5.44PALWPRR503 pKa = 11.84KK504 pKa = 8.67RR505 pKa = 11.84RR506 pKa = 11.84RR507 pKa = 11.84LDD509 pKa = 3.23LFF511 pKa = 4.04
Molecular weight: 55.38 kDa
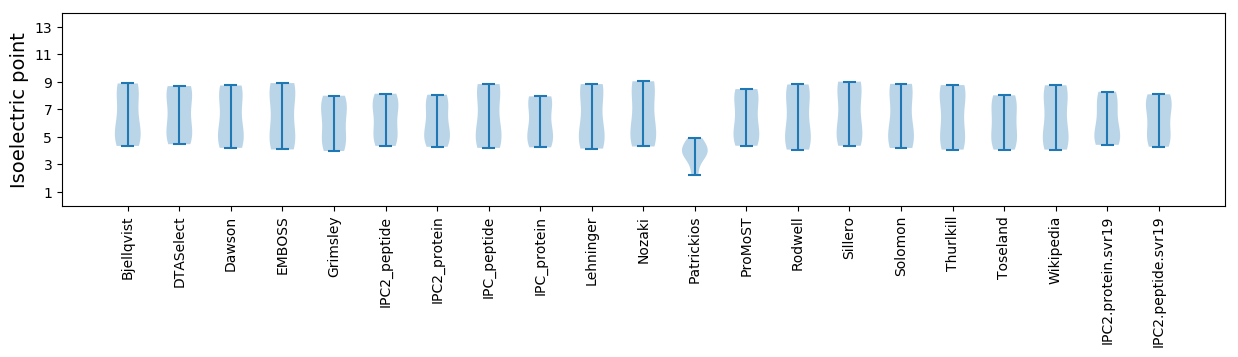
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|E7BQ87|E7BQ87_9PAPI Protein E7 OS=Human papillomavirus type 131 OX=909330 GN=E7 PE=3 SV=1
MM1 pKa = 7.95ASTYY5 pKa = 10.57PNNLVDD11 pKa = 5.3LCSACNISFFNLHH24 pKa = 6.86LDD26 pKa = 4.43CLFCKK31 pKa = 10.47HH32 pKa = 6.48KK33 pKa = 10.33LTILDD38 pKa = 3.74LAGFHH43 pKa = 5.84QKK45 pKa = 9.69QLSVVWRR52 pKa = 11.84NDD54 pKa = 2.96KK55 pKa = 11.36AFGSCSKK62 pKa = 10.55CLRR65 pKa = 11.84LTANYY70 pKa = 9.42EE71 pKa = 3.69RR72 pKa = 11.84EE73 pKa = 3.7NYY75 pKa = 8.75YY76 pKa = 10.3RR77 pKa = 11.84CSVKK81 pKa = 10.47GSLISGLLKK90 pKa = 10.21TPLKK94 pKa = 10.6DD95 pKa = 2.89ITVRR99 pKa = 11.84CLYY102 pKa = 10.37CFQLLDD108 pKa = 5.0LIEE111 pKa = 4.45KK112 pKa = 7.22TQHH115 pKa = 6.15HH116 pKa = 6.55LGDD119 pKa = 3.67RR120 pKa = 11.84DD121 pKa = 3.52FHH123 pKa = 6.7LVRR126 pKa = 11.84NHH128 pKa = 5.51WRR130 pKa = 11.84GCCRR134 pKa = 11.84NCYY137 pKa = 10.11SYY139 pKa = 11.33EE140 pKa = 4.0GG141 pKa = 3.63
MM1 pKa = 7.95ASTYY5 pKa = 10.57PNNLVDD11 pKa = 5.3LCSACNISFFNLHH24 pKa = 6.86LDD26 pKa = 4.43CLFCKK31 pKa = 10.47HH32 pKa = 6.48KK33 pKa = 10.33LTILDD38 pKa = 3.74LAGFHH43 pKa = 5.84QKK45 pKa = 9.69QLSVVWRR52 pKa = 11.84NDD54 pKa = 2.96KK55 pKa = 11.36AFGSCSKK62 pKa = 10.55CLRR65 pKa = 11.84LTANYY70 pKa = 9.42EE71 pKa = 3.69RR72 pKa = 11.84EE73 pKa = 3.7NYY75 pKa = 8.75YY76 pKa = 10.3RR77 pKa = 11.84CSVKK81 pKa = 10.47GSLISGLLKK90 pKa = 10.21TPLKK94 pKa = 10.6DD95 pKa = 2.89ITVRR99 pKa = 11.84CLYY102 pKa = 10.37CFQLLDD108 pKa = 5.0LIEE111 pKa = 4.45KK112 pKa = 7.22TQHH115 pKa = 6.15HH116 pKa = 6.55LGDD119 pKa = 3.67RR120 pKa = 11.84DD121 pKa = 3.52FHH123 pKa = 6.7LVRR126 pKa = 11.84NHH128 pKa = 5.51WRR130 pKa = 11.84GCCRR134 pKa = 11.84NCYY137 pKa = 10.11SYY139 pKa = 11.33EE140 pKa = 4.0GG141 pKa = 3.63
Molecular weight: 16.39 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2447 |
94 |
663 |
349.6 |
39.27 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.885 ± 0.529 | 2.411 ± 0.779 |
6.334 ± 0.447 | 6.253 ± 0.617 |
4.986 ± 0.663 | 5.599 ± 0.661 |
2.166 ± 0.359 | 5.149 ± 0.731 |
5.353 ± 0.852 | 10.012 ± 0.817 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.512 ± 0.42 | 5.313 ± 0.282 |
5.885 ± 1.025 | 3.269 ± 0.416 |
4.904 ± 0.688 | 7.601 ± 0.579 |
6.457 ± 0.742 | 6.498 ± 0.403 |
1.389 ± 0.228 | 3.024 ± 0.467 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
