
Lynx canadensis faeces associated genomovirus CL1 58
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; unclassified Genomoviridae
Average proteome isoelectric point is 7.2
Get precalculated fractions of proteins
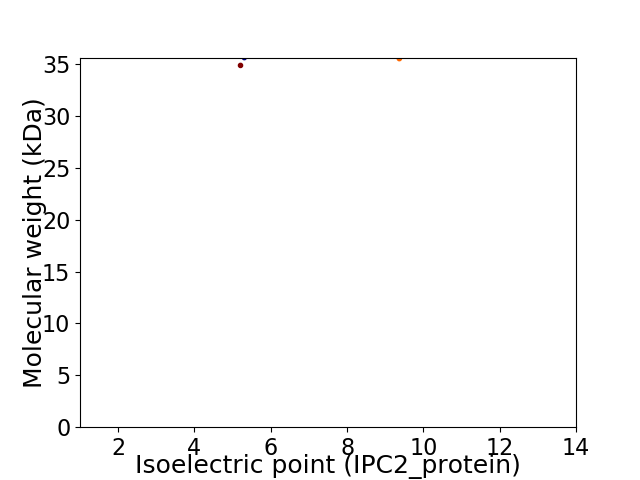
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A2Z5CI18|A0A2Z5CI18_9VIRU Replication-associated protein OS=Lynx canadensis faeces associated genomovirus CL1 58 OX=2219126 PE=3 SV=1
MM1 pKa = 7.75PSFVANFKK9 pKa = 10.24YY10 pKa = 10.83ALVTYY15 pKa = 6.91SQCGDD20 pKa = 3.79LDD22 pKa = 3.55PWSVMEE28 pKa = 4.86LFSSLGAEE36 pKa = 4.24CIIGRR41 pKa = 11.84EE42 pKa = 4.01HH43 pKa = 7.75HH44 pKa = 6.83EE45 pKa = 4.72DD46 pKa = 3.59GGTHH50 pKa = 5.89LHH52 pKa = 5.88VFVDD56 pKa = 4.55FGRR59 pKa = 11.84KK60 pKa = 7.93FRR62 pKa = 11.84SRR64 pKa = 11.84ATNVFDD70 pKa = 5.08VDD72 pKa = 3.5GHH74 pKa = 6.44HH75 pKa = 7.29PNIEE79 pKa = 4.06PSKK82 pKa = 8.78GTPEE86 pKa = 4.01KK87 pKa = 11.01GFDD90 pKa = 3.46YY91 pKa = 10.65AIKK94 pKa = 10.85DD95 pKa = 3.45GDD97 pKa = 4.33VVCGGLGRR105 pKa = 11.84PEE107 pKa = 4.06PSRR110 pKa = 11.84GGNSGVLAKK119 pKa = 7.87WTRR122 pKa = 11.84ITGATDD128 pKa = 3.13KK129 pKa = 11.52QEE131 pKa = 4.11FWDD134 pKa = 4.65LVHH137 pKa = 6.98EE138 pKa = 5.1LDD140 pKa = 4.6PKK142 pKa = 10.76SAACSFTSLSKK153 pKa = 10.06YY154 pKa = 10.56ADD156 pKa = 2.96WKK158 pKa = 10.68FAVDD162 pKa = 4.22PPCYY166 pKa = 8.96EE167 pKa = 4.02HH168 pKa = 7.56PGGFDD173 pKa = 3.41FSDD176 pKa = 3.9GSFDD180 pKa = 4.32GRR182 pKa = 11.84SEE184 pKa = 4.2CKK186 pKa = 10.23SLVMYY191 pKa = 10.37GPSRR195 pKa = 11.84TGKK198 pKa = 8.21TMWARR203 pKa = 11.84SLGAHH208 pKa = 7.19LYY210 pKa = 10.52CVGLLSGDD218 pKa = 3.43EE219 pKa = 4.17CMKK222 pKa = 10.97AEE224 pKa = 4.07YY225 pKa = 10.43ADD227 pKa = 3.75YY228 pKa = 11.08AIFDD232 pKa = 4.25DD233 pKa = 4.69LRR235 pKa = 11.84GGVKK239 pKa = 9.8FFPAFKK245 pKa = 9.49EE246 pKa = 3.89WLGAQQYY253 pKa = 7.12VTVKK257 pKa = 10.5RR258 pKa = 11.84LYY260 pKa = 10.56HH261 pKa = 5.72EE262 pKa = 4.69PKK264 pKa = 9.56LVKK267 pKa = 9.05WGKK270 pKa = 9.69PSIFLTNSDD279 pKa = 4.19PRR281 pKa = 11.84DD282 pKa = 3.48EE283 pKa = 4.34ATGDD287 pKa = 3.5DD288 pKa = 4.13RR289 pKa = 11.84DD290 pKa = 3.78WLEE293 pKa = 4.08RR294 pKa = 11.84NCDD297 pKa = 4.42FIYY300 pKa = 9.55VQEE303 pKa = 5.02PIFHH307 pKa = 7.21ANTEE311 pKa = 4.14
MM1 pKa = 7.75PSFVANFKK9 pKa = 10.24YY10 pKa = 10.83ALVTYY15 pKa = 6.91SQCGDD20 pKa = 3.79LDD22 pKa = 3.55PWSVMEE28 pKa = 4.86LFSSLGAEE36 pKa = 4.24CIIGRR41 pKa = 11.84EE42 pKa = 4.01HH43 pKa = 7.75HH44 pKa = 6.83EE45 pKa = 4.72DD46 pKa = 3.59GGTHH50 pKa = 5.89LHH52 pKa = 5.88VFVDD56 pKa = 4.55FGRR59 pKa = 11.84KK60 pKa = 7.93FRR62 pKa = 11.84SRR64 pKa = 11.84ATNVFDD70 pKa = 5.08VDD72 pKa = 3.5GHH74 pKa = 6.44HH75 pKa = 7.29PNIEE79 pKa = 4.06PSKK82 pKa = 8.78GTPEE86 pKa = 4.01KK87 pKa = 11.01GFDD90 pKa = 3.46YY91 pKa = 10.65AIKK94 pKa = 10.85DD95 pKa = 3.45GDD97 pKa = 4.33VVCGGLGRR105 pKa = 11.84PEE107 pKa = 4.06PSRR110 pKa = 11.84GGNSGVLAKK119 pKa = 7.87WTRR122 pKa = 11.84ITGATDD128 pKa = 3.13KK129 pKa = 11.52QEE131 pKa = 4.11FWDD134 pKa = 4.65LVHH137 pKa = 6.98EE138 pKa = 5.1LDD140 pKa = 4.6PKK142 pKa = 10.76SAACSFTSLSKK153 pKa = 10.06YY154 pKa = 10.56ADD156 pKa = 2.96WKK158 pKa = 10.68FAVDD162 pKa = 4.22PPCYY166 pKa = 8.96EE167 pKa = 4.02HH168 pKa = 7.56PGGFDD173 pKa = 3.41FSDD176 pKa = 3.9GSFDD180 pKa = 4.32GRR182 pKa = 11.84SEE184 pKa = 4.2CKK186 pKa = 10.23SLVMYY191 pKa = 10.37GPSRR195 pKa = 11.84TGKK198 pKa = 8.21TMWARR203 pKa = 11.84SLGAHH208 pKa = 7.19LYY210 pKa = 10.52CVGLLSGDD218 pKa = 3.43EE219 pKa = 4.17CMKK222 pKa = 10.97AEE224 pKa = 4.07YY225 pKa = 10.43ADD227 pKa = 3.75YY228 pKa = 11.08AIFDD232 pKa = 4.25DD233 pKa = 4.69LRR235 pKa = 11.84GGVKK239 pKa = 9.8FFPAFKK245 pKa = 9.49EE246 pKa = 3.89WLGAQQYY253 pKa = 7.12VTVKK257 pKa = 10.5RR258 pKa = 11.84LYY260 pKa = 10.56HH261 pKa = 5.72EE262 pKa = 4.69PKK264 pKa = 9.56LVKK267 pKa = 9.05WGKK270 pKa = 9.69PSIFLTNSDD279 pKa = 4.19PRR281 pKa = 11.84DD282 pKa = 3.48EE283 pKa = 4.34ATGDD287 pKa = 3.5DD288 pKa = 4.13RR289 pKa = 11.84DD290 pKa = 3.78WLEE293 pKa = 4.08RR294 pKa = 11.84NCDD297 pKa = 4.42FIYY300 pKa = 9.55VQEE303 pKa = 5.02PIFHH307 pKa = 7.21ANTEE311 pKa = 4.14
Molecular weight: 34.88 kDa
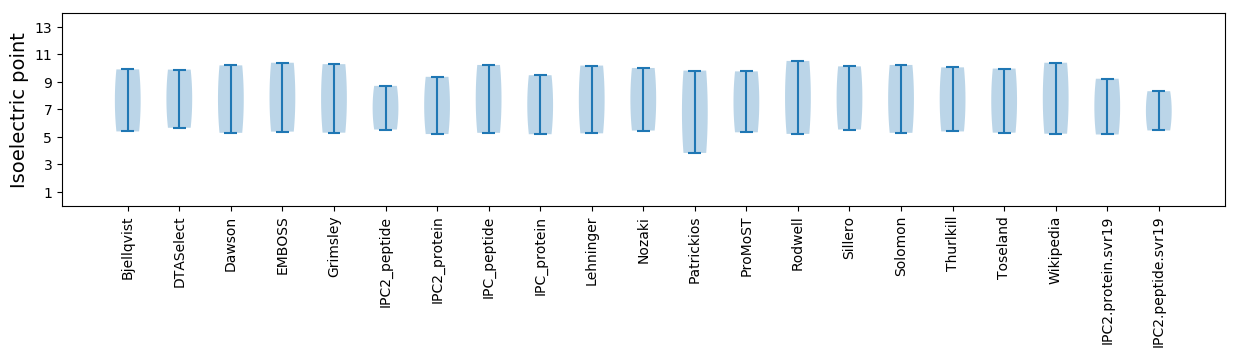
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A2Z5CI18|A0A2Z5CI18_9VIRU Replication-associated protein OS=Lynx canadensis faeces associated genomovirus CL1 58 OX=2219126 PE=3 SV=1
MM1 pKa = 7.8PARR4 pKa = 11.84AYY6 pKa = 9.86RR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.34SGSYY13 pKa = 9.45RR14 pKa = 11.84GRR16 pKa = 11.84KK17 pKa = 9.06RR18 pKa = 11.84GGPTKK23 pKa = 10.37RR24 pKa = 11.84AWSNYY29 pKa = 8.11KK30 pKa = 10.19RR31 pKa = 11.84STGRR35 pKa = 11.84TRR37 pKa = 11.84PYY39 pKa = 8.19RR40 pKa = 11.84TSRR43 pKa = 11.84TRR45 pKa = 11.84QKK47 pKa = 10.64PMSRR51 pKa = 11.84KK52 pKa = 9.74KK53 pKa = 10.01ILNVTSEE60 pKa = 4.03KK61 pKa = 10.67KK62 pKa = 9.49RR63 pKa = 11.84DD64 pKa = 4.56KK65 pKa = 10.14MLCWTNTIAGTPQGGTTYY83 pKa = 8.82TQSPAILTGAGADD96 pKa = 3.47PYY98 pKa = 11.28LFAWCATARR107 pKa = 11.84DD108 pKa = 4.19NTTSTGGAAHH118 pKa = 7.57AGTKK122 pKa = 9.38FDD124 pKa = 3.59KK125 pKa = 10.21ATRR128 pKa = 11.84TASEE132 pKa = 4.82CYY134 pKa = 8.93MVGLSEE140 pKa = 5.4RR141 pKa = 11.84IEE143 pKa = 4.04IQCADD148 pKa = 3.69GLPWQWRR155 pKa = 11.84RR156 pKa = 11.84ICFTMKK162 pKa = 10.51GGVGLDD168 pKa = 3.5GTLTTGSTFSSYY180 pKa = 11.46SEE182 pKa = 4.15TTSGYY187 pKa = 9.17VRR189 pKa = 11.84TVNSVLLTDD198 pKa = 3.86RR199 pKa = 11.84TSLYY203 pKa = 11.13SLLFAGAQSSDD214 pKa = 2.6WSDD217 pKa = 3.44PMVAPLDD224 pKa = 3.53RR225 pKa = 11.84RR226 pKa = 11.84RR227 pKa = 11.84INVKK231 pKa = 9.65YY232 pKa = 10.89DD233 pKa = 2.98KK234 pKa = 8.37TTSIASGNEE243 pKa = 3.47DD244 pKa = 3.28GVIRR248 pKa = 11.84KK249 pKa = 5.55YY250 pKa = 9.38TKK252 pKa = 7.92WHH254 pKa = 6.36PMGKK258 pKa = 7.54TLMYY262 pKa = 10.87DD263 pKa = 3.38DD264 pKa = 5.72DD265 pKa = 5.0EE266 pKa = 4.65NTGAEE271 pKa = 4.18NPAVISVDD279 pKa = 3.2NKK281 pKa = 10.65QGMGDD286 pKa = 3.57FWVLDD291 pKa = 4.45FIKK294 pKa = 10.35PRR296 pKa = 11.84MGSTSSNQLSFRR308 pKa = 11.84PEE310 pKa = 3.66STLYY314 pKa = 8.15WHH316 pKa = 6.78EE317 pKa = 4.04RR318 pKa = 3.41
MM1 pKa = 7.8PARR4 pKa = 11.84AYY6 pKa = 9.86RR7 pKa = 11.84RR8 pKa = 11.84KK9 pKa = 9.34SGSYY13 pKa = 9.45RR14 pKa = 11.84GRR16 pKa = 11.84KK17 pKa = 9.06RR18 pKa = 11.84GGPTKK23 pKa = 10.37RR24 pKa = 11.84AWSNYY29 pKa = 8.11KK30 pKa = 10.19RR31 pKa = 11.84STGRR35 pKa = 11.84TRR37 pKa = 11.84PYY39 pKa = 8.19RR40 pKa = 11.84TSRR43 pKa = 11.84TRR45 pKa = 11.84QKK47 pKa = 10.64PMSRR51 pKa = 11.84KK52 pKa = 9.74KK53 pKa = 10.01ILNVTSEE60 pKa = 4.03KK61 pKa = 10.67KK62 pKa = 9.49RR63 pKa = 11.84DD64 pKa = 4.56KK65 pKa = 10.14MLCWTNTIAGTPQGGTTYY83 pKa = 8.82TQSPAILTGAGADD96 pKa = 3.47PYY98 pKa = 11.28LFAWCATARR107 pKa = 11.84DD108 pKa = 4.19NTTSTGGAAHH118 pKa = 7.57AGTKK122 pKa = 9.38FDD124 pKa = 3.59KK125 pKa = 10.21ATRR128 pKa = 11.84TASEE132 pKa = 4.82CYY134 pKa = 8.93MVGLSEE140 pKa = 5.4RR141 pKa = 11.84IEE143 pKa = 4.04IQCADD148 pKa = 3.69GLPWQWRR155 pKa = 11.84RR156 pKa = 11.84ICFTMKK162 pKa = 10.51GGVGLDD168 pKa = 3.5GTLTTGSTFSSYY180 pKa = 11.46SEE182 pKa = 4.15TTSGYY187 pKa = 9.17VRR189 pKa = 11.84TVNSVLLTDD198 pKa = 3.86RR199 pKa = 11.84TSLYY203 pKa = 11.13SLLFAGAQSSDD214 pKa = 2.6WSDD217 pKa = 3.44PMVAPLDD224 pKa = 3.53RR225 pKa = 11.84RR226 pKa = 11.84RR227 pKa = 11.84INVKK231 pKa = 9.65YY232 pKa = 10.89DD233 pKa = 2.98KK234 pKa = 8.37TTSIASGNEE243 pKa = 3.47DD244 pKa = 3.28GVIRR248 pKa = 11.84KK249 pKa = 5.55YY250 pKa = 9.38TKK252 pKa = 7.92WHH254 pKa = 6.36PMGKK258 pKa = 7.54TLMYY262 pKa = 10.87DD263 pKa = 3.38DD264 pKa = 5.72DD265 pKa = 5.0EE266 pKa = 4.65NTGAEE271 pKa = 4.18NPAVISVDD279 pKa = 3.2NKK281 pKa = 10.65QGMGDD286 pKa = 3.57FWVLDD291 pKa = 4.45FIKK294 pKa = 10.35PRR296 pKa = 11.84MGSTSSNQLSFRR308 pKa = 11.84PEE310 pKa = 3.66STLYY314 pKa = 8.15WHH316 pKa = 6.78EE317 pKa = 4.04RR318 pKa = 3.41
Molecular weight: 35.53 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
629 |
311 |
318 |
314.5 |
35.21 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.677 ± 0.172 | 2.226 ± 0.466 |
7.154 ± 1.066 | 4.61 ± 1.046 |
4.61 ± 1.495 | 9.539 ± 0.299 |
2.226 ± 0.915 | 3.18 ± 0.199 |
6.2 ± 0.064 | 6.2 ± 0.385 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.385 ± 0.542 | 2.862 ± 0.426 |
4.928 ± 0.375 | 2.067 ± 0.32 |
6.995 ± 1.516 | 8.267 ± 0.833 |
8.108 ± 2.517 | 4.928 ± 0.824 |
2.703 ± 0.091 | 4.134 ± 0.192 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
