
Ecklonia radiata-associated virus 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.66
Get precalculated fractions of proteins
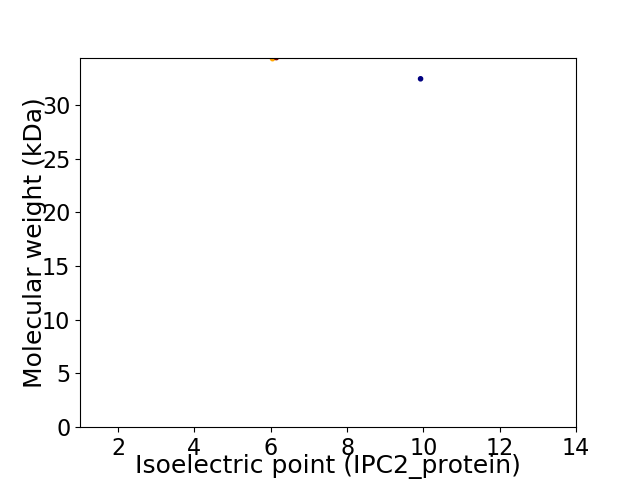
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A3G2CHT1|A0A3G2CHT1_9CIRC Rep-A OS=Ecklonia radiata-associated virus 5 OX=2480195 PE=4 SV=1
MM1 pKa = 7.45SVPSSTRR8 pKa = 11.84GKK10 pKa = 9.93RR11 pKa = 11.84WVFTLNNHH19 pKa = 5.76TEE21 pKa = 4.25GEE23 pKa = 4.36VQLLSDD29 pKa = 5.04LLTSEE34 pKa = 4.28HH35 pKa = 5.71VSYY38 pKa = 10.99GIFGRR43 pKa = 11.84EE44 pKa = 3.95VGEE47 pKa = 4.05SGTPHH52 pKa = 6.02LQGYY56 pKa = 9.26CIFADD61 pKa = 3.73RR62 pKa = 11.84KK63 pKa = 10.34RR64 pKa = 11.84FNQVRR69 pKa = 11.84ALFGXRR75 pKa = 11.84YY76 pKa = 8.88HH77 pKa = 8.42VEE79 pKa = 3.61ISRR82 pKa = 11.84GTPAQASQYY91 pKa = 9.62CKK93 pKa = 10.25KK94 pKa = 10.78DD95 pKa = 2.95GDD97 pKa = 3.85FEE99 pKa = 4.65EE100 pKa = 5.61FGEE103 pKa = 4.89FPGTQGKK110 pKa = 10.12RR111 pKa = 11.84SDD113 pKa = 3.65WEE115 pKa = 4.42SLKK118 pKa = 10.68LWCEE122 pKa = 3.93TQTRR126 pKa = 11.84QPTDD130 pKa = 3.17LEE132 pKa = 4.16LFYY135 pKa = 11.03AFPSLFGRR143 pKa = 11.84YY144 pKa = 7.03EE145 pKa = 3.78KK146 pKa = 10.51SVRR149 pKa = 11.84KK150 pKa = 9.6ICNLCIKK157 pKa = 10.07CDD159 pKa = 3.6PXXIGEE165 pKa = 3.93PRR167 pKa = 11.84GWQRR171 pKa = 11.84DD172 pKa = 3.63XEE174 pKa = 4.31EE175 pKa = 4.99RR176 pKa = 11.84LLDD179 pKa = 4.29EE180 pKa = 5.42PDD182 pKa = 3.54DD183 pKa = 4.04RR184 pKa = 11.84CIEE187 pKa = 3.96FVLDD191 pKa = 3.54YY192 pKa = 10.34EE193 pKa = 4.72GNSGKK198 pKa = 10.09SWFVRR203 pKa = 11.84YY204 pKa = 10.29YY205 pKa = 9.02MLKK208 pKa = 9.54HH209 pKa = 5.8QGSAQMLSVGKK220 pKa = 10.1RR221 pKa = 11.84DD222 pKa = 5.39DD223 pKa = 3.38IAHH226 pKa = 6.4CXDD229 pKa = 3.01VSRR232 pKa = 11.84RR233 pKa = 11.84VFFFDD238 pKa = 3.18IPRR241 pKa = 11.84GGMEE245 pKa = 4.14FLQYY249 pKa = 10.89GALEE253 pKa = 4.72GIKK256 pKa = 10.44NGFIFSPKK264 pKa = 9.74YY265 pKa = 10.1EE266 pKa = 4.26STTKK270 pKa = 10.58VLLNKK275 pKa = 9.86AHH277 pKa = 6.52VVVLCNEE284 pKa = 4.4DD285 pKa = 3.69PDD287 pKa = 4.11MLKK290 pKa = 10.88LSLDD294 pKa = 3.46RR295 pKa = 11.84YY296 pKa = 10.62KK297 pKa = 9.77ITNLXXLL304 pKa = 5.31
MM1 pKa = 7.45SVPSSTRR8 pKa = 11.84GKK10 pKa = 9.93RR11 pKa = 11.84WVFTLNNHH19 pKa = 5.76TEE21 pKa = 4.25GEE23 pKa = 4.36VQLLSDD29 pKa = 5.04LLTSEE34 pKa = 4.28HH35 pKa = 5.71VSYY38 pKa = 10.99GIFGRR43 pKa = 11.84EE44 pKa = 3.95VGEE47 pKa = 4.05SGTPHH52 pKa = 6.02LQGYY56 pKa = 9.26CIFADD61 pKa = 3.73RR62 pKa = 11.84KK63 pKa = 10.34RR64 pKa = 11.84FNQVRR69 pKa = 11.84ALFGXRR75 pKa = 11.84YY76 pKa = 8.88HH77 pKa = 8.42VEE79 pKa = 3.61ISRR82 pKa = 11.84GTPAQASQYY91 pKa = 9.62CKK93 pKa = 10.25KK94 pKa = 10.78DD95 pKa = 2.95GDD97 pKa = 3.85FEE99 pKa = 4.65EE100 pKa = 5.61FGEE103 pKa = 4.89FPGTQGKK110 pKa = 10.12RR111 pKa = 11.84SDD113 pKa = 3.65WEE115 pKa = 4.42SLKK118 pKa = 10.68LWCEE122 pKa = 3.93TQTRR126 pKa = 11.84QPTDD130 pKa = 3.17LEE132 pKa = 4.16LFYY135 pKa = 11.03AFPSLFGRR143 pKa = 11.84YY144 pKa = 7.03EE145 pKa = 3.78KK146 pKa = 10.51SVRR149 pKa = 11.84KK150 pKa = 9.6ICNLCIKK157 pKa = 10.07CDD159 pKa = 3.6PXXIGEE165 pKa = 3.93PRR167 pKa = 11.84GWQRR171 pKa = 11.84DD172 pKa = 3.63XEE174 pKa = 4.31EE175 pKa = 4.99RR176 pKa = 11.84LLDD179 pKa = 4.29EE180 pKa = 5.42PDD182 pKa = 3.54DD183 pKa = 4.04RR184 pKa = 11.84CIEE187 pKa = 3.96FVLDD191 pKa = 3.54YY192 pKa = 10.34EE193 pKa = 4.72GNSGKK198 pKa = 10.09SWFVRR203 pKa = 11.84YY204 pKa = 10.29YY205 pKa = 9.02MLKK208 pKa = 9.54HH209 pKa = 5.8QGSAQMLSVGKK220 pKa = 10.1RR221 pKa = 11.84DD222 pKa = 5.39DD223 pKa = 3.38IAHH226 pKa = 6.4CXDD229 pKa = 3.01VSRR232 pKa = 11.84RR233 pKa = 11.84VFFFDD238 pKa = 3.18IPRR241 pKa = 11.84GGMEE245 pKa = 4.14FLQYY249 pKa = 10.89GALEE253 pKa = 4.72GIKK256 pKa = 10.44NGFIFSPKK264 pKa = 9.74YY265 pKa = 10.1EE266 pKa = 4.26STTKK270 pKa = 10.58VLLNKK275 pKa = 9.86AHH277 pKa = 6.52VVVLCNEE284 pKa = 4.4DD285 pKa = 3.69PDD287 pKa = 4.11MLKK290 pKa = 10.88LSLDD294 pKa = 3.46RR295 pKa = 11.84YY296 pKa = 10.62KK297 pKa = 9.77ITNLXXLL304 pKa = 5.31
Molecular weight: 34.3 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3G2CHT1|A0A3G2CHT1_9CIRC Rep-A OS=Ecklonia radiata-associated virus 5 OX=2480195 PE=4 SV=1
MM1 pKa = 7.93AIRR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 10.17RR7 pKa = 11.84IILPLHH13 pKa = 6.16RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84VRR18 pKa = 11.84PRR20 pKa = 11.84LYY22 pKa = 8.93KK23 pKa = 10.28TKK25 pKa = 9.06TRR27 pKa = 11.84TYY29 pKa = 9.95RR30 pKa = 11.84RR31 pKa = 11.84SYY33 pKa = 9.04GNKK36 pKa = 9.05RR37 pKa = 11.84MRR39 pKa = 11.84RR40 pKa = 11.84MQDD43 pKa = 3.05FSPRR47 pKa = 11.84AIANPVGLSNAKK59 pKa = 8.57TRR61 pKa = 11.84LLTFSQDD68 pKa = 3.18SPDD71 pKa = 3.58RR72 pKa = 11.84AFVPYY77 pKa = 10.4IMTNIPFTAANAPDD91 pKa = 3.39SRR93 pKa = 11.84QRR95 pKa = 11.84NIINIRR101 pKa = 11.84GFKK104 pKa = 9.35IQVVFQNTQRR114 pKa = 11.84PAFYY118 pKa = 10.68LNWAVIHH125 pKa = 6.51PKK127 pKa = 10.69LSIDD131 pKa = 3.65DD132 pKa = 4.28TSVSTQDD139 pKa = 3.7FFRR142 pKa = 11.84TYY144 pKa = 9.82NANRR148 pKa = 11.84AVDD151 pKa = 5.6LITDD155 pKa = 3.46NTGVVNNIAQDD166 pKa = 3.79VPTHH170 pKa = 5.77GMSSINTDD178 pKa = 2.21KK179 pKa = 10.91WVVLKK184 pKa = 10.54HH185 pKa = 6.04KK186 pKa = 9.37RR187 pKa = 11.84TLLAGAINNPDD198 pKa = 3.49DD199 pKa = 3.65NALXFXATNCQIPKK213 pKa = 9.65GNRR216 pKa = 11.84VVIDD220 pKa = 3.85KK221 pKa = 9.38YY222 pKa = 11.18IKK224 pKa = 9.45MGRR227 pKa = 11.84QFRR230 pKa = 11.84FPNDD234 pKa = 3.22DD235 pKa = 3.31STFPEE240 pKa = 4.67GGNTIFCYY248 pKa = 10.14WLTNLNNYY256 pKa = 9.69LGSQNTSXDD265 pKa = 3.31VSVIRR270 pKa = 11.84TTGRR274 pKa = 11.84IITYY278 pKa = 9.72FRR280 pKa = 11.84EE281 pKa = 4.34TKK283 pKa = 10.27NN284 pKa = 3.15
MM1 pKa = 7.93AIRR4 pKa = 11.84RR5 pKa = 11.84KK6 pKa = 10.17RR7 pKa = 11.84IILPLHH13 pKa = 6.16RR14 pKa = 11.84RR15 pKa = 11.84RR16 pKa = 11.84VRR18 pKa = 11.84PRR20 pKa = 11.84LYY22 pKa = 8.93KK23 pKa = 10.28TKK25 pKa = 9.06TRR27 pKa = 11.84TYY29 pKa = 9.95RR30 pKa = 11.84RR31 pKa = 11.84SYY33 pKa = 9.04GNKK36 pKa = 9.05RR37 pKa = 11.84MRR39 pKa = 11.84RR40 pKa = 11.84MQDD43 pKa = 3.05FSPRR47 pKa = 11.84AIANPVGLSNAKK59 pKa = 8.57TRR61 pKa = 11.84LLTFSQDD68 pKa = 3.18SPDD71 pKa = 3.58RR72 pKa = 11.84AFVPYY77 pKa = 10.4IMTNIPFTAANAPDD91 pKa = 3.39SRR93 pKa = 11.84QRR95 pKa = 11.84NIINIRR101 pKa = 11.84GFKK104 pKa = 9.35IQVVFQNTQRR114 pKa = 11.84PAFYY118 pKa = 10.68LNWAVIHH125 pKa = 6.51PKK127 pKa = 10.69LSIDD131 pKa = 3.65DD132 pKa = 4.28TSVSTQDD139 pKa = 3.7FFRR142 pKa = 11.84TYY144 pKa = 9.82NANRR148 pKa = 11.84AVDD151 pKa = 5.6LITDD155 pKa = 3.46NTGVVNNIAQDD166 pKa = 3.79VPTHH170 pKa = 5.77GMSSINTDD178 pKa = 2.21KK179 pKa = 10.91WVVLKK184 pKa = 10.54HH185 pKa = 6.04KK186 pKa = 9.37RR187 pKa = 11.84TLLAGAINNPDD198 pKa = 3.49DD199 pKa = 3.65NALXFXATNCQIPKK213 pKa = 9.65GNRR216 pKa = 11.84VVIDD220 pKa = 3.85KK221 pKa = 9.38YY222 pKa = 11.18IKK224 pKa = 9.45MGRR227 pKa = 11.84QFRR230 pKa = 11.84FPNDD234 pKa = 3.22DD235 pKa = 3.31STFPEE240 pKa = 4.67GGNTIFCYY248 pKa = 10.14WLTNLNNYY256 pKa = 9.69LGSQNTSXDD265 pKa = 3.31VSVIRR270 pKa = 11.84TTGRR274 pKa = 11.84IITYY278 pKa = 9.72FRR280 pKa = 11.84EE281 pKa = 4.34TKK283 pKa = 10.27NN284 pKa = 3.15
Molecular weight: 32.43 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
588 |
284 |
304 |
294.0 |
33.36 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.422 ± 1.0 | 1.871 ± 0.746 |
6.122 ± 0.087 | 4.252 ± 2.268 |
5.782 ± 0.32 | 6.293 ± 1.321 |
1.871 ± 0.296 | 5.952 ± 1.372 |
5.442 ± 0.328 | 7.483 ± 1.182 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.871 ± 0.155 | 6.122 ± 2.164 |
4.592 ± 0.441 | 3.912 ± 0.025 |
8.844 ± 1.099 | 6.122 ± 0.537 |
6.463 ± 1.496 | 5.782 ± 0.13 |
1.361 ± 0.194 | 3.741 ± 0.141 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
