
Avon-Heathcote Estuary associated circular virus 21
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 7.4
Get precalculated fractions of proteins
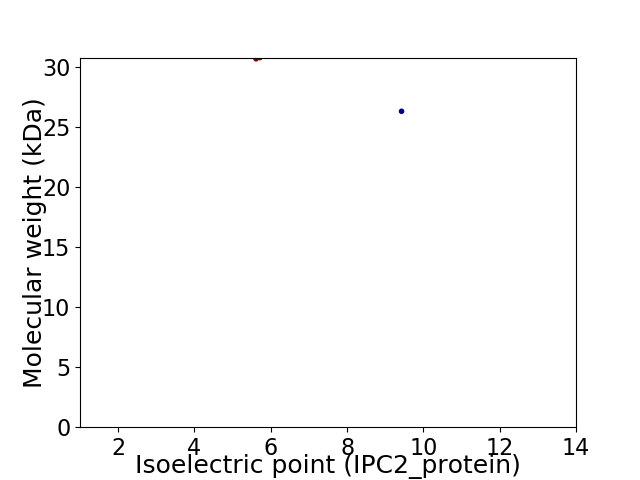
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IMK1|A0A0C5IMK1_9CIRC Replication-associated protein OS=Avon-Heathcote Estuary associated circular virus 21 OX=1618245 PE=4 SV=1
MM1 pKa = 7.56PKK3 pKa = 10.09ARR5 pKa = 11.84PSNGSRR11 pKa = 11.84FWCFTINNPTDD22 pKa = 4.3DD23 pKa = 5.79DD24 pKa = 4.16NQQLADD30 pKa = 4.15LMSNDD35 pKa = 3.39LCTYY39 pKa = 9.55LCYY42 pKa = 10.55GRR44 pKa = 11.84EE45 pKa = 4.18TGEE48 pKa = 4.29SEE50 pKa = 4.25TPHH53 pKa = 4.99YY54 pKa = 9.51QAYY57 pKa = 9.83IEE59 pKa = 4.61LLVPNDD65 pKa = 3.58FLGLRR70 pKa = 11.84NDD72 pKa = 3.7LKK74 pKa = 11.17EE75 pKa = 5.02LILNHH80 pKa = 6.58ASEE83 pKa = 4.33AAPRR87 pKa = 11.84QEE89 pKa = 3.87TTASRR94 pKa = 11.84KK95 pKa = 9.5ISTQSNMVSGSPIAKK110 pKa = 9.39GKK112 pKa = 10.79GMTSRR117 pKa = 11.84FFTEE121 pKa = 3.85YY122 pKa = 10.58RR123 pKa = 11.84AMHH126 pKa = 6.03AMEE129 pKa = 5.48RR130 pKa = 11.84DD131 pKa = 3.74APTKK135 pKa = 10.14VYY137 pKa = 10.57VFHH140 pKa = 7.26GPTASGKK147 pKa = 7.74TRR149 pKa = 11.84AAFDD153 pKa = 4.1MNCHH157 pKa = 5.16QMDD160 pKa = 4.66YY161 pKa = 10.3IAPFFTDD168 pKa = 3.59PQNAPVLLFDD178 pKa = 5.67DD179 pKa = 3.51VHH181 pKa = 7.76NPVNLFGRR189 pKa = 11.84RR190 pKa = 11.84LFLRR194 pKa = 11.84ITDD197 pKa = 4.37RR198 pKa = 11.84YY199 pKa = 7.94PMKK202 pKa = 10.46VRR204 pKa = 11.84CLGKK208 pKa = 9.8YY209 pKa = 8.62VEE211 pKa = 4.57WNPTVIIFTTNDD223 pKa = 4.3DD224 pKa = 4.12PNTWNLDD231 pKa = 3.38AACRR235 pKa = 11.84RR236 pKa = 11.84RR237 pKa = 11.84IDD239 pKa = 3.91EE240 pKa = 4.9IIAFPLDD247 pKa = 3.46KK248 pKa = 11.08GGLDD252 pKa = 3.92ALPACTQDD260 pKa = 3.16QAKK263 pKa = 10.46PGDD266 pKa = 3.87LPQCNN271 pKa = 3.48
MM1 pKa = 7.56PKK3 pKa = 10.09ARR5 pKa = 11.84PSNGSRR11 pKa = 11.84FWCFTINNPTDD22 pKa = 4.3DD23 pKa = 5.79DD24 pKa = 4.16NQQLADD30 pKa = 4.15LMSNDD35 pKa = 3.39LCTYY39 pKa = 9.55LCYY42 pKa = 10.55GRR44 pKa = 11.84EE45 pKa = 4.18TGEE48 pKa = 4.29SEE50 pKa = 4.25TPHH53 pKa = 4.99YY54 pKa = 9.51QAYY57 pKa = 9.83IEE59 pKa = 4.61LLVPNDD65 pKa = 3.58FLGLRR70 pKa = 11.84NDD72 pKa = 3.7LKK74 pKa = 11.17EE75 pKa = 5.02LILNHH80 pKa = 6.58ASEE83 pKa = 4.33AAPRR87 pKa = 11.84QEE89 pKa = 3.87TTASRR94 pKa = 11.84KK95 pKa = 9.5ISTQSNMVSGSPIAKK110 pKa = 9.39GKK112 pKa = 10.79GMTSRR117 pKa = 11.84FFTEE121 pKa = 3.85YY122 pKa = 10.58RR123 pKa = 11.84AMHH126 pKa = 6.03AMEE129 pKa = 5.48RR130 pKa = 11.84DD131 pKa = 3.74APTKK135 pKa = 10.14VYY137 pKa = 10.57VFHH140 pKa = 7.26GPTASGKK147 pKa = 7.74TRR149 pKa = 11.84AAFDD153 pKa = 4.1MNCHH157 pKa = 5.16QMDD160 pKa = 4.66YY161 pKa = 10.3IAPFFTDD168 pKa = 3.59PQNAPVLLFDD178 pKa = 5.67DD179 pKa = 3.51VHH181 pKa = 7.76NPVNLFGRR189 pKa = 11.84RR190 pKa = 11.84LFLRR194 pKa = 11.84ITDD197 pKa = 4.37RR198 pKa = 11.84YY199 pKa = 7.94PMKK202 pKa = 10.46VRR204 pKa = 11.84CLGKK208 pKa = 9.8YY209 pKa = 8.62VEE211 pKa = 4.57WNPTVIIFTTNDD223 pKa = 4.3DD224 pKa = 4.12PNTWNLDD231 pKa = 3.38AACRR235 pKa = 11.84RR236 pKa = 11.84RR237 pKa = 11.84IDD239 pKa = 3.91EE240 pKa = 4.9IIAFPLDD247 pKa = 3.46KK248 pKa = 11.08GGLDD252 pKa = 3.92ALPACTQDD260 pKa = 3.16QAKK263 pKa = 10.46PGDD266 pKa = 3.87LPQCNN271 pKa = 3.48
Molecular weight: 30.65 kDa
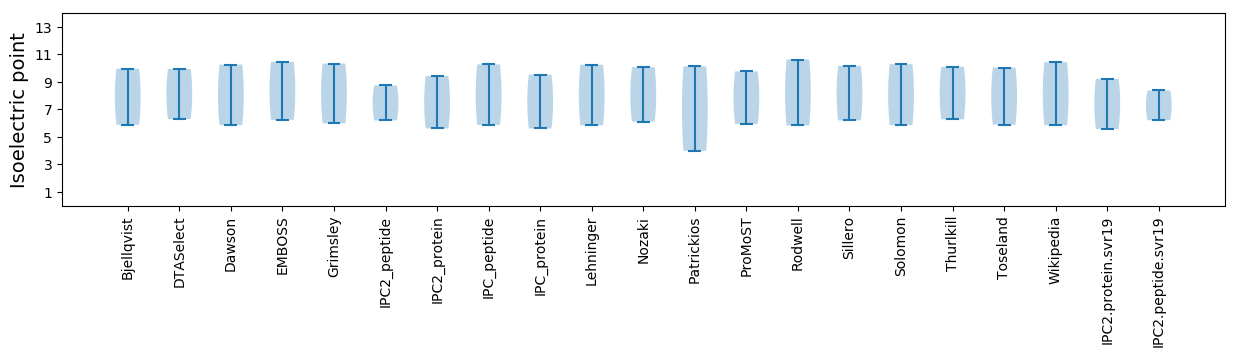
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IMK1|A0A0C5IMK1_9CIRC Replication-associated protein OS=Avon-Heathcote Estuary associated circular virus 21 OX=1618245 PE=4 SV=1
MM1 pKa = 7.84PKK3 pKa = 9.99RR4 pKa = 11.84GRR6 pKa = 11.84SRR8 pKa = 11.84TRR10 pKa = 11.84KK11 pKa = 8.48KK12 pKa = 10.69RR13 pKa = 11.84PWQRR17 pKa = 11.84RR18 pKa = 11.84SSSGSRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 8.02PLRR30 pKa = 11.84SRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84PLALKK39 pKa = 9.54QHH41 pKa = 6.65AFCEE45 pKa = 4.23RR46 pKa = 11.84AEE48 pKa = 4.04NEE50 pKa = 3.62QTIVINTEE58 pKa = 3.44AAATGLFEE66 pKa = 4.62TFTLSKK72 pKa = 9.74MRR74 pKa = 11.84QQASYY79 pKa = 9.76AQIFEE84 pKa = 4.44FYY86 pKa = 10.56RR87 pKa = 11.84IDD89 pKa = 3.52KK90 pKa = 10.36VVATFRR96 pKa = 11.84YY97 pKa = 10.08KK98 pKa = 10.61NVNNATEE105 pKa = 4.39TTGRR109 pKa = 11.84AQNEE113 pKa = 4.51VNPLLYY119 pKa = 10.58FKK121 pKa = 10.82VDD123 pKa = 3.58HH124 pKa = 7.05NDD126 pKa = 3.23NTSDD130 pKa = 3.98SLSTMKK136 pKa = 10.89DD137 pKa = 2.92SMKK140 pKa = 10.44TKK142 pKa = 8.92THH144 pKa = 6.05MFTNNNPEE152 pKa = 3.88FSIQLKK158 pKa = 9.62PAIQSEE164 pKa = 4.86MYY166 pKa = 8.89RR167 pKa = 11.84TTLTAAYY174 pKa = 7.15TPKK177 pKa = 9.05WGQWIPCDD185 pKa = 4.13DD186 pKa = 4.09PTVPHH191 pKa = 6.62YY192 pKa = 11.23GMKK195 pKa = 10.2AYY197 pKa = 10.67CCAFKK202 pKa = 10.93NADD205 pKa = 3.68WNPGSLQVSFKK216 pKa = 10.99YY217 pKa = 10.57YY218 pKa = 10.92VSFKK222 pKa = 11.05NNEE225 pKa = 3.71
MM1 pKa = 7.84PKK3 pKa = 9.99RR4 pKa = 11.84GRR6 pKa = 11.84SRR8 pKa = 11.84TRR10 pKa = 11.84KK11 pKa = 8.48KK12 pKa = 10.69RR13 pKa = 11.84PWQRR17 pKa = 11.84RR18 pKa = 11.84SSSGSRR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84YY27 pKa = 8.02PLRR30 pKa = 11.84SRR32 pKa = 11.84RR33 pKa = 11.84RR34 pKa = 11.84PLALKK39 pKa = 9.54QHH41 pKa = 6.65AFCEE45 pKa = 4.23RR46 pKa = 11.84AEE48 pKa = 4.04NEE50 pKa = 3.62QTIVINTEE58 pKa = 3.44AAATGLFEE66 pKa = 4.62TFTLSKK72 pKa = 9.74MRR74 pKa = 11.84QQASYY79 pKa = 9.76AQIFEE84 pKa = 4.44FYY86 pKa = 10.56RR87 pKa = 11.84IDD89 pKa = 3.52KK90 pKa = 10.36VVATFRR96 pKa = 11.84YY97 pKa = 10.08KK98 pKa = 10.61NVNNATEE105 pKa = 4.39TTGRR109 pKa = 11.84AQNEE113 pKa = 4.51VNPLLYY119 pKa = 10.58FKK121 pKa = 10.82VDD123 pKa = 3.58HH124 pKa = 7.05NDD126 pKa = 3.23NTSDD130 pKa = 3.98SLSTMKK136 pKa = 10.89DD137 pKa = 2.92SMKK140 pKa = 10.44TKK142 pKa = 8.92THH144 pKa = 6.05MFTNNNPEE152 pKa = 3.88FSIQLKK158 pKa = 9.62PAIQSEE164 pKa = 4.86MYY166 pKa = 8.89RR167 pKa = 11.84TTLTAAYY174 pKa = 7.15TPKK177 pKa = 9.05WGQWIPCDD185 pKa = 4.13DD186 pKa = 4.09PTVPHH191 pKa = 6.62YY192 pKa = 11.23GMKK195 pKa = 10.2AYY197 pKa = 10.67CCAFKK202 pKa = 10.93NADD205 pKa = 3.68WNPGSLQVSFKK216 pKa = 10.99YY217 pKa = 10.57YY218 pKa = 10.92VSFKK222 pKa = 11.05NNEE225 pKa = 3.71
Molecular weight: 26.29 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
496 |
225 |
271 |
248.0 |
28.47 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.863 ± 0.193 | 2.419 ± 0.403 |
6.048 ± 1.566 | 4.435 ± 0.285 |
5.242 ± 0.057 | 4.234 ± 0.705 |
2.016 ± 0.15 | 3.831 ± 0.452 |
5.645 ± 1.2 | 6.653 ± 1.108 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.226 ± 0.072 | 6.855 ± 0.161 |
6.452 ± 0.702 | 4.234 ± 0.411 |
7.661 ± 0.771 | 5.847 ± 1.073 |
8.065 ± 0.518 | 3.831 ± 0.106 |
1.411 ± 0.23 | 4.032 ± 0.538 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
