
Neorhizobium sp. S3-V5DH
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Hyphomicrobiales; Rhizobiaceae; Rhizobium/Agrobacterium group; Neorhizobium; unclassified Neorhizobium
Average proteome isoelectric point is 6.48
Get precalculated fractions of proteins
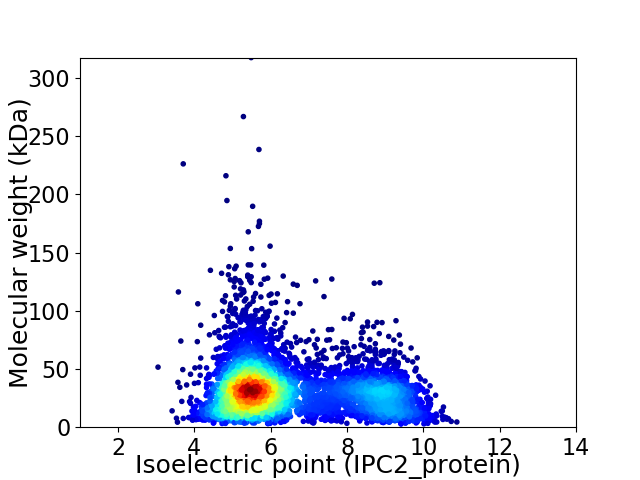
Virtual 2D-PAGE plot for 5607 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4R3XFL1|A0A4R3XFL1_9RHIZ DeoR family transcriptional regulator OS=Neorhizobium sp. S3-V5DH OX=2485166 GN=EDE09_10649 PE=4 SV=1
MM1 pKa = 7.24TAMDD5 pKa = 5.66KK6 pKa = 9.45ITEE9 pKa = 4.53EE10 pKa = 3.76IAHH13 pKa = 6.7FIGLFHH19 pKa = 6.12TTVEE23 pKa = 4.29EE24 pKa = 3.81ARR26 pKa = 11.84QRR28 pKa = 11.84DD29 pKa = 3.76AYY31 pKa = 11.35LDD33 pKa = 3.56FSYY36 pKa = 9.8YY37 pKa = 6.59TQKK40 pKa = 11.25DD41 pKa = 3.62HH42 pKa = 7.03LHH44 pKa = 5.95TEE46 pKa = 4.25PFISSTFDD54 pKa = 2.9APYY57 pKa = 10.34EE58 pKa = 3.97FLGFDD63 pKa = 3.48PDD65 pKa = 3.67VFYY68 pKa = 10.98KK69 pKa = 10.8APAYY73 pKa = 9.65LRR75 pKa = 11.84PLEE78 pKa = 4.63HH79 pKa = 7.27PSFASWQHH87 pKa = 5.79HH88 pKa = 5.67PPILVVRR95 pKa = 11.84DD96 pKa = 3.9DD97 pKa = 4.03SPYY100 pKa = 10.01FPPEE104 pKa = 3.63QTEE107 pKa = 4.3EE108 pKa = 4.15ILPGARR114 pKa = 11.84IGQAFTVPPIEE125 pKa = 4.6PPGSVVNYY133 pKa = 9.66IIQAASLSDD142 pKa = 4.17DD143 pKa = 3.44DD144 pKa = 4.29TFNVGGSNLEE154 pKa = 3.95FHH156 pKa = 7.64PDD158 pKa = 3.64PVNDD162 pKa = 3.79AALLKK167 pKa = 10.75AADD170 pKa = 4.81DD171 pKa = 4.35VLSHH175 pKa = 6.72APIGHH180 pKa = 7.09PEE182 pKa = 4.06MPGSSAEE189 pKa = 4.38LIDD192 pKa = 4.59VINDD196 pKa = 2.89IAFQLKK202 pKa = 10.36ALVAAPGSPMEE213 pKa = 4.24TFIEE217 pKa = 4.23HH218 pKa = 6.08SPVIEE223 pKa = 3.81GVYY226 pKa = 10.77VNGEE230 pKa = 4.14LVEE233 pKa = 4.54DD234 pKa = 4.11APKK237 pKa = 10.99LEE239 pKa = 5.43DD240 pKa = 3.62YY241 pKa = 11.01FSFDD245 pKa = 3.52DD246 pKa = 4.41EE247 pKa = 4.63EE248 pKa = 4.27NAEE251 pKa = 4.23EE252 pKa = 4.56NDD254 pKa = 3.67AEE256 pKa = 4.52SPADD260 pKa = 3.77EE261 pKa = 5.81DD262 pKa = 4.98ADD264 pKa = 3.87NDD266 pKa = 4.2DD267 pKa = 5.21AVGADD272 pKa = 3.74ADD274 pKa = 4.58EE275 pKa = 5.86DD276 pKa = 4.33DD277 pKa = 4.95GPPPNVVISEE287 pKa = 4.56DD288 pKa = 3.53GSITIDD294 pKa = 3.37NSVEE298 pKa = 4.18LIAGDD303 pKa = 3.5NTVVNQAVVKK313 pKa = 10.02NLWTGATVTAVVGDD327 pKa = 3.91HH328 pKa = 6.3VEE330 pKa = 3.89INAIVQTNAIWDD342 pKa = 3.87TDD344 pKa = 4.53SITSAIDD351 pKa = 3.08GWNSEE356 pKa = 4.32DD357 pKa = 4.71SPNEE361 pKa = 3.74LFNIATFEE369 pKa = 4.06RR370 pKa = 11.84TDD372 pKa = 3.63PLEE375 pKa = 4.51EE376 pKa = 4.65GSGDD380 pKa = 3.28QAAAGGFPSAWAITEE395 pKa = 3.94IQGDD399 pKa = 3.98LLITNWLEE407 pKa = 3.83QYY409 pKa = 11.58VFMTDD414 pKa = 2.77NDD416 pKa = 3.75VGILSSSGVTTSVIAGDD433 pKa = 3.69NTSVNQTSIFEE444 pKa = 4.64LGFSFDD450 pKa = 4.69LIIVGGSLYY459 pKa = 10.48DD460 pKa = 3.59ANIIHH465 pKa = 5.38QTNVLFDD472 pKa = 4.03NDD474 pKa = 3.36VVGAVSGFEE483 pKa = 4.07TTGQGSIGSSGNLLWNQAHH502 pKa = 7.36IYY504 pKa = 9.7NVGGADD510 pKa = 3.49RR511 pKa = 11.84FGEE514 pKa = 4.3LPSAYY519 pKa = 10.17LDD521 pKa = 3.88AANDD525 pKa = 3.6MAAGGRR531 pKa = 11.84DD532 pKa = 3.44LSKK535 pKa = 11.25GVLTDD540 pKa = 3.46PAFAGLGGLRR550 pKa = 11.84VLYY553 pKa = 10.44ISGDD557 pKa = 3.64FLNIQYY563 pKa = 10.31ISQTNIVGDD572 pKa = 3.75SDD574 pKa = 4.42QIALAMNALDD584 pKa = 4.44PQTDD588 pKa = 3.82AAWTVSTGANALINNAAILDD608 pKa = 3.92FDD610 pKa = 4.67SFGHH614 pKa = 6.04TYY616 pKa = 10.76VGGQQYY622 pKa = 10.38SEE624 pKa = 4.02EE625 pKa = 4.23TLFQAEE631 pKa = 5.14LISHH635 pKa = 5.91QPDD638 pKa = 4.5LLTGDD643 pKa = 4.43PDD645 pKa = 4.03TLVNEE650 pKa = 4.34AVVFLDD656 pKa = 5.31DD657 pKa = 4.83SMLDD661 pKa = 3.68GEE663 pKa = 4.49FEE665 pKa = 4.42EE666 pKa = 4.95MGVTPAEE673 pKa = 4.07QDD675 pKa = 3.03NGHH678 pKa = 5.66YY679 pKa = 10.61QNDD682 pKa = 4.18GLQHH686 pKa = 5.34VLGG689 pKa = 4.52
MM1 pKa = 7.24TAMDD5 pKa = 5.66KK6 pKa = 9.45ITEE9 pKa = 4.53EE10 pKa = 3.76IAHH13 pKa = 6.7FIGLFHH19 pKa = 6.12TTVEE23 pKa = 4.29EE24 pKa = 3.81ARR26 pKa = 11.84QRR28 pKa = 11.84DD29 pKa = 3.76AYY31 pKa = 11.35LDD33 pKa = 3.56FSYY36 pKa = 9.8YY37 pKa = 6.59TQKK40 pKa = 11.25DD41 pKa = 3.62HH42 pKa = 7.03LHH44 pKa = 5.95TEE46 pKa = 4.25PFISSTFDD54 pKa = 2.9APYY57 pKa = 10.34EE58 pKa = 3.97FLGFDD63 pKa = 3.48PDD65 pKa = 3.67VFYY68 pKa = 10.98KK69 pKa = 10.8APAYY73 pKa = 9.65LRR75 pKa = 11.84PLEE78 pKa = 4.63HH79 pKa = 7.27PSFASWQHH87 pKa = 5.79HH88 pKa = 5.67PPILVVRR95 pKa = 11.84DD96 pKa = 3.9DD97 pKa = 4.03SPYY100 pKa = 10.01FPPEE104 pKa = 3.63QTEE107 pKa = 4.3EE108 pKa = 4.15ILPGARR114 pKa = 11.84IGQAFTVPPIEE125 pKa = 4.6PPGSVVNYY133 pKa = 9.66IIQAASLSDD142 pKa = 4.17DD143 pKa = 3.44DD144 pKa = 4.29TFNVGGSNLEE154 pKa = 3.95FHH156 pKa = 7.64PDD158 pKa = 3.64PVNDD162 pKa = 3.79AALLKK167 pKa = 10.75AADD170 pKa = 4.81DD171 pKa = 4.35VLSHH175 pKa = 6.72APIGHH180 pKa = 7.09PEE182 pKa = 4.06MPGSSAEE189 pKa = 4.38LIDD192 pKa = 4.59VINDD196 pKa = 2.89IAFQLKK202 pKa = 10.36ALVAAPGSPMEE213 pKa = 4.24TFIEE217 pKa = 4.23HH218 pKa = 6.08SPVIEE223 pKa = 3.81GVYY226 pKa = 10.77VNGEE230 pKa = 4.14LVEE233 pKa = 4.54DD234 pKa = 4.11APKK237 pKa = 10.99LEE239 pKa = 5.43DD240 pKa = 3.62YY241 pKa = 11.01FSFDD245 pKa = 3.52DD246 pKa = 4.41EE247 pKa = 4.63EE248 pKa = 4.27NAEE251 pKa = 4.23EE252 pKa = 4.56NDD254 pKa = 3.67AEE256 pKa = 4.52SPADD260 pKa = 3.77EE261 pKa = 5.81DD262 pKa = 4.98ADD264 pKa = 3.87NDD266 pKa = 4.2DD267 pKa = 5.21AVGADD272 pKa = 3.74ADD274 pKa = 4.58EE275 pKa = 5.86DD276 pKa = 4.33DD277 pKa = 4.95GPPPNVVISEE287 pKa = 4.56DD288 pKa = 3.53GSITIDD294 pKa = 3.37NSVEE298 pKa = 4.18LIAGDD303 pKa = 3.5NTVVNQAVVKK313 pKa = 10.02NLWTGATVTAVVGDD327 pKa = 3.91HH328 pKa = 6.3VEE330 pKa = 3.89INAIVQTNAIWDD342 pKa = 3.87TDD344 pKa = 4.53SITSAIDD351 pKa = 3.08GWNSEE356 pKa = 4.32DD357 pKa = 4.71SPNEE361 pKa = 3.74LFNIATFEE369 pKa = 4.06RR370 pKa = 11.84TDD372 pKa = 3.63PLEE375 pKa = 4.51EE376 pKa = 4.65GSGDD380 pKa = 3.28QAAAGGFPSAWAITEE395 pKa = 3.94IQGDD399 pKa = 3.98LLITNWLEE407 pKa = 3.83QYY409 pKa = 11.58VFMTDD414 pKa = 2.77NDD416 pKa = 3.75VGILSSSGVTTSVIAGDD433 pKa = 3.69NTSVNQTSIFEE444 pKa = 4.64LGFSFDD450 pKa = 4.69LIIVGGSLYY459 pKa = 10.48DD460 pKa = 3.59ANIIHH465 pKa = 5.38QTNVLFDD472 pKa = 4.03NDD474 pKa = 3.36VVGAVSGFEE483 pKa = 4.07TTGQGSIGSSGNLLWNQAHH502 pKa = 7.36IYY504 pKa = 9.7NVGGADD510 pKa = 3.49RR511 pKa = 11.84FGEE514 pKa = 4.3LPSAYY519 pKa = 10.17LDD521 pKa = 3.88AANDD525 pKa = 3.6MAAGGRR531 pKa = 11.84DD532 pKa = 3.44LSKK535 pKa = 11.25GVLTDD540 pKa = 3.46PAFAGLGGLRR550 pKa = 11.84VLYY553 pKa = 10.44ISGDD557 pKa = 3.64FLNIQYY563 pKa = 10.31ISQTNIVGDD572 pKa = 3.75SDD574 pKa = 4.42QIALAMNALDD584 pKa = 4.44PQTDD588 pKa = 3.82AAWTVSTGANALINNAAILDD608 pKa = 3.92FDD610 pKa = 4.67SFGHH614 pKa = 6.04TYY616 pKa = 10.76VGGQQYY622 pKa = 10.38SEE624 pKa = 4.02EE625 pKa = 4.23TLFQAEE631 pKa = 5.14LISHH635 pKa = 5.91QPDD638 pKa = 4.5LLTGDD643 pKa = 4.43PDD645 pKa = 4.03TLVNEE650 pKa = 4.34AVVFLDD656 pKa = 5.31DD657 pKa = 4.83SMLDD661 pKa = 3.68GEE663 pKa = 4.49FEE665 pKa = 4.42EE666 pKa = 4.95MGVTPAEE673 pKa = 4.07QDD675 pKa = 3.03NGHH678 pKa = 5.66YY679 pKa = 10.61QNDD682 pKa = 4.18GLQHH686 pKa = 5.34VLGG689 pKa = 4.52
Molecular weight: 74.04 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4R3XBB3|A0A4R3XBB3_9RHIZ 2-keto-4-pentenoate hydratase OS=Neorhizobium sp. S3-V5DH OX=2485166 GN=EDE09_11018 PE=4 SV=1
MM1 pKa = 7.32RR2 pKa = 11.84QLAKK6 pKa = 10.35FLSATGRR13 pKa = 11.84RR14 pKa = 11.84LRR16 pKa = 11.84TLGKK20 pKa = 9.81VVSHH24 pKa = 6.69FFRR27 pKa = 11.84KK28 pKa = 9.77GKK30 pKa = 10.34LGLNIAGGVVTT41 pKa = 5.52
MM1 pKa = 7.32RR2 pKa = 11.84QLAKK6 pKa = 10.35FLSATGRR13 pKa = 11.84RR14 pKa = 11.84LRR16 pKa = 11.84TLGKK20 pKa = 9.81VVSHH24 pKa = 6.69FFRR27 pKa = 11.84KK28 pKa = 9.77GKK30 pKa = 10.34LGLNIAGGVVTT41 pKa = 5.52
Molecular weight: 4.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1719071 |
25 |
2831 |
306.6 |
33.45 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.603 ± 0.045 | 0.798 ± 0.008 |
5.561 ± 0.027 | 5.995 ± 0.032 |
3.982 ± 0.02 | 8.321 ± 0.028 |
2.022 ± 0.015 | 5.72 ± 0.027 |
3.774 ± 0.026 | 9.977 ± 0.041 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.638 ± 0.015 | 2.852 ± 0.018 |
4.972 ± 0.025 | 3.096 ± 0.019 |
6.741 ± 0.029 | 5.783 ± 0.023 |
5.23 ± 0.019 | 7.288 ± 0.026 |
1.313 ± 0.014 | 2.333 ± 0.017 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
