
Monascus purpureus (Red mold) (Monascus anka)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Fungi; Dikarya; Ascomycota; saccharomyceta; Pezizomycotina; leotiomyceta; Eurotiomycetes; Eurotiomycetidae; Eurotiales; Aspergillaceae; Monascus
Average proteome isoelectric point is 6.53
Get precalculated fractions of proteins
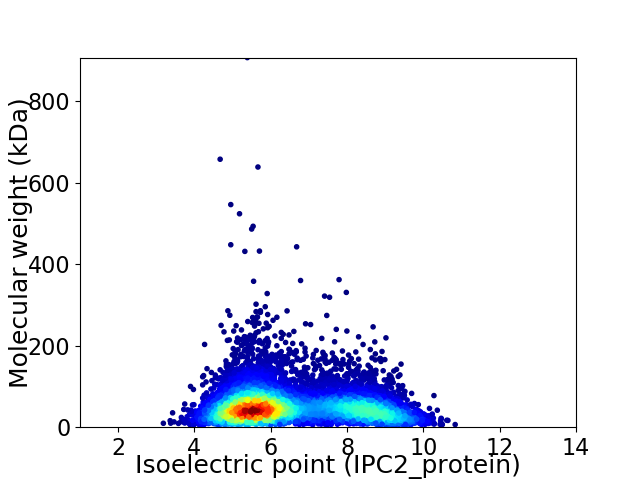
Virtual 2D-PAGE plot for 8080 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions

Protein with the lowest isoelectric point:
>tr|A0A507QS44|A0A507QS44_MONPU Amidase (Fragment) OS=Monascus purpureus OX=5098 GN=MPDQ_000374 PE=3 SV=1
MM1 pKa = 7.75PPLAHH6 pKa = 7.06LLSAFLRR13 pKa = 11.84IAEE16 pKa = 4.17ISFAAIVAGIIGNFLYY32 pKa = 10.51EE33 pKa = 4.27FEE35 pKa = 5.24LFSGDD40 pKa = 3.31WIDD43 pKa = 4.43ARR45 pKa = 11.84WIYY48 pKa = 10.7TEE50 pKa = 4.26VIAGISIVLGLILVIPFATSFFAWPLDD77 pKa = 3.33ILIALSWFAAFGILVDD93 pKa = 3.7GAEE96 pKa = 5.01RR97 pKa = 11.84LDD99 pKa = 4.49CDD101 pKa = 4.15GNPYY105 pKa = 9.55WVIFLGDD112 pKa = 3.95SFCDD116 pKa = 3.0EE117 pKa = 4.07WKK119 pKa = 10.59AAEE122 pKa = 4.26AFCFLSGVAWIVSGFVVSSPLGQSITGLVVSDD154 pKa = 5.03KK155 pKa = 10.99II156 pKa = 4.05
MM1 pKa = 7.75PPLAHH6 pKa = 7.06LLSAFLRR13 pKa = 11.84IAEE16 pKa = 4.17ISFAAIVAGIIGNFLYY32 pKa = 10.51EE33 pKa = 4.27FEE35 pKa = 5.24LFSGDD40 pKa = 3.31WIDD43 pKa = 4.43ARR45 pKa = 11.84WIYY48 pKa = 10.7TEE50 pKa = 4.26VIAGISIVLGLILVIPFATSFFAWPLDD77 pKa = 3.33ILIALSWFAAFGILVDD93 pKa = 3.7GAEE96 pKa = 5.01RR97 pKa = 11.84LDD99 pKa = 4.49CDD101 pKa = 4.15GNPYY105 pKa = 9.55WVIFLGDD112 pKa = 3.95SFCDD116 pKa = 3.0EE117 pKa = 4.07WKK119 pKa = 10.59AAEE122 pKa = 4.26AFCFLSGVAWIVSGFVVSSPLGQSITGLVVSDD154 pKa = 5.03KK155 pKa = 10.99II156 pKa = 4.05
Molecular weight: 17.03 kDa
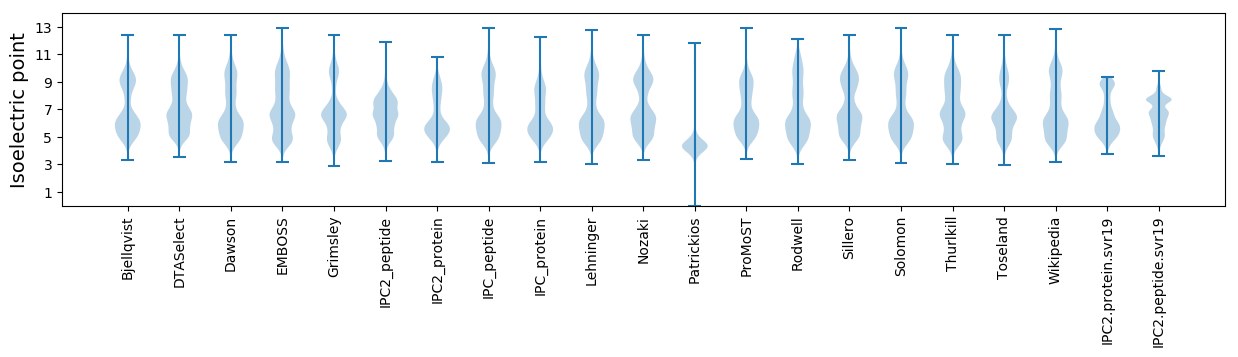
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A507R2I8|A0A507R2I8_MONPU CHY-type domain-containing protein OS=Monascus purpureus OX=5098 GN=MPDQ_007514 PE=4 SV=1
MM1 pKa = 7.8RR2 pKa = 11.84GAIAPIPGDD11 pKa = 3.44GKK13 pKa = 10.42IDD15 pKa = 3.46IRR17 pKa = 11.84ILKK20 pKa = 9.56PPLRR24 pKa = 11.84AFAVGYY30 pKa = 10.16LSSTAPRR37 pKa = 11.84LVSYY41 pKa = 7.49LTRR44 pKa = 11.84SRR46 pKa = 11.84KK47 pKa = 9.89NKK49 pKa = 9.1VGQKK53 pKa = 9.93RR54 pKa = 11.84RR55 pKa = 11.84FEE57 pKa = 4.17EE58 pKa = 4.08LAEE61 pKa = 4.14LLIKK65 pKa = 9.51DD66 pKa = 3.95TRR68 pKa = 11.84FHH70 pKa = 7.01AFPSFLALLVGGSTIIPVLAQRR92 pKa = 11.84LAASIATRR100 pKa = 11.84FRR102 pKa = 11.84LRR104 pKa = 11.84RR105 pKa = 11.84DD106 pKa = 3.59VITSSAFVRR115 pKa = 11.84LTRR118 pKa = 11.84FISALLSAWFCFRR131 pKa = 11.84ILNRR135 pKa = 11.84RR136 pKa = 11.84HH137 pKa = 6.61DD138 pKa = 3.98RR139 pKa = 11.84LQRR142 pKa = 11.84HH143 pKa = 5.69YY144 pKa = 10.99GGQIEE149 pKa = 4.6DD150 pKa = 3.57ASEE153 pKa = 4.05KK154 pKa = 10.58QSASDD159 pKa = 3.47PTDD162 pKa = 3.62SIEE165 pKa = 3.77QRR167 pKa = 11.84QRR169 pKa = 11.84RR170 pKa = 11.84SPEE173 pKa = 3.83FAGSTLDD180 pKa = 3.55LTVLVATRR188 pKa = 11.84AADD191 pKa = 3.53AVACIGWARR200 pKa = 11.84WRR202 pKa = 11.84AWRR205 pKa = 11.84MTRR208 pKa = 11.84NRR210 pKa = 11.84WTLAEE215 pKa = 4.14SVAPKK220 pKa = 10.5VADD223 pKa = 3.48AGIFVLSSAVIMWAWFYY240 pKa = 11.26LPEE243 pKa = 4.67RR244 pKa = 11.84LPRR247 pKa = 11.84SYY249 pKa = 11.26GRR251 pKa = 11.84WIDD254 pKa = 3.38EE255 pKa = 4.07AAQVDD260 pKa = 3.58ARR262 pKa = 11.84LIEE265 pKa = 4.05VLRR268 pKa = 11.84RR269 pKa = 11.84ARR271 pKa = 11.84RR272 pKa = 11.84GEE274 pKa = 3.82FVYY277 pKa = 10.39GKK279 pKa = 8.74DD280 pKa = 2.95TGQAPFLQTMCKK292 pKa = 10.18DD293 pKa = 3.88YY294 pKa = 10.58GWPMVWGDD302 pKa = 3.54PAKK305 pKa = 9.98TIPLPCEE312 pKa = 4.11VVHH315 pKa = 6.36MGYY318 pKa = 10.76GPNCEE323 pKa = 3.63IYY325 pKa = 10.24AARR328 pKa = 11.84RR329 pKa = 11.84FLKK332 pKa = 9.69TFKK335 pKa = 10.64FGFWTYY341 pKa = 10.68FPLQAALRR349 pKa = 11.84ARR351 pKa = 11.84GLRR354 pKa = 11.84SMRR357 pKa = 11.84TLVRR361 pKa = 11.84IAFDD365 pKa = 3.62ASRR368 pKa = 11.84SSAFLALFVTLFYY381 pKa = 11.35YY382 pKa = 8.94SVCLARR388 pKa = 11.84TRR390 pKa = 11.84LGPKK394 pKa = 9.87VFDD397 pKa = 4.27KK398 pKa = 11.57NKK400 pKa = 7.1VTPMMWDD407 pKa = 2.87SGLCVGAGCLMCGWSVLVEE426 pKa = 3.86KK427 pKa = 10.69ASRR430 pKa = 11.84RR431 pKa = 11.84RR432 pKa = 11.84EE433 pKa = 3.7LSLFVAPRR441 pKa = 11.84AAATLLPRR449 pKa = 11.84VYY451 pKa = 10.58DD452 pKa = 3.03KK453 pKa = 11.04QYY455 pKa = 9.13QHH457 pKa = 7.02RR458 pKa = 11.84EE459 pKa = 3.72RR460 pKa = 11.84IAFAVCAAILSTCLQEE476 pKa = 4.21RR477 pKa = 11.84PEE479 pKa = 4.0MVRR482 pKa = 11.84GFFRR486 pKa = 11.84KK487 pKa = 9.73IGARR491 pKa = 11.84LIAA494 pKa = 4.7
MM1 pKa = 7.8RR2 pKa = 11.84GAIAPIPGDD11 pKa = 3.44GKK13 pKa = 10.42IDD15 pKa = 3.46IRR17 pKa = 11.84ILKK20 pKa = 9.56PPLRR24 pKa = 11.84AFAVGYY30 pKa = 10.16LSSTAPRR37 pKa = 11.84LVSYY41 pKa = 7.49LTRR44 pKa = 11.84SRR46 pKa = 11.84KK47 pKa = 9.89NKK49 pKa = 9.1VGQKK53 pKa = 9.93RR54 pKa = 11.84RR55 pKa = 11.84FEE57 pKa = 4.17EE58 pKa = 4.08LAEE61 pKa = 4.14LLIKK65 pKa = 9.51DD66 pKa = 3.95TRR68 pKa = 11.84FHH70 pKa = 7.01AFPSFLALLVGGSTIIPVLAQRR92 pKa = 11.84LAASIATRR100 pKa = 11.84FRR102 pKa = 11.84LRR104 pKa = 11.84RR105 pKa = 11.84DD106 pKa = 3.59VITSSAFVRR115 pKa = 11.84LTRR118 pKa = 11.84FISALLSAWFCFRR131 pKa = 11.84ILNRR135 pKa = 11.84RR136 pKa = 11.84HH137 pKa = 6.61DD138 pKa = 3.98RR139 pKa = 11.84LQRR142 pKa = 11.84HH143 pKa = 5.69YY144 pKa = 10.99GGQIEE149 pKa = 4.6DD150 pKa = 3.57ASEE153 pKa = 4.05KK154 pKa = 10.58QSASDD159 pKa = 3.47PTDD162 pKa = 3.62SIEE165 pKa = 3.77QRR167 pKa = 11.84QRR169 pKa = 11.84RR170 pKa = 11.84SPEE173 pKa = 3.83FAGSTLDD180 pKa = 3.55LTVLVATRR188 pKa = 11.84AADD191 pKa = 3.53AVACIGWARR200 pKa = 11.84WRR202 pKa = 11.84AWRR205 pKa = 11.84MTRR208 pKa = 11.84NRR210 pKa = 11.84WTLAEE215 pKa = 4.14SVAPKK220 pKa = 10.5VADD223 pKa = 3.48AGIFVLSSAVIMWAWFYY240 pKa = 11.26LPEE243 pKa = 4.67RR244 pKa = 11.84LPRR247 pKa = 11.84SYY249 pKa = 11.26GRR251 pKa = 11.84WIDD254 pKa = 3.38EE255 pKa = 4.07AAQVDD260 pKa = 3.58ARR262 pKa = 11.84LIEE265 pKa = 4.05VLRR268 pKa = 11.84RR269 pKa = 11.84ARR271 pKa = 11.84RR272 pKa = 11.84GEE274 pKa = 3.82FVYY277 pKa = 10.39GKK279 pKa = 8.74DD280 pKa = 2.95TGQAPFLQTMCKK292 pKa = 10.18DD293 pKa = 3.88YY294 pKa = 10.58GWPMVWGDD302 pKa = 3.54PAKK305 pKa = 9.98TIPLPCEE312 pKa = 4.11VVHH315 pKa = 6.36MGYY318 pKa = 10.76GPNCEE323 pKa = 3.63IYY325 pKa = 10.24AARR328 pKa = 11.84RR329 pKa = 11.84FLKK332 pKa = 9.69TFKK335 pKa = 10.64FGFWTYY341 pKa = 10.68FPLQAALRR349 pKa = 11.84ARR351 pKa = 11.84GLRR354 pKa = 11.84SMRR357 pKa = 11.84TLVRR361 pKa = 11.84IAFDD365 pKa = 3.62ASRR368 pKa = 11.84SSAFLALFVTLFYY381 pKa = 11.35YY382 pKa = 8.94SVCLARR388 pKa = 11.84TRR390 pKa = 11.84LGPKK394 pKa = 9.87VFDD397 pKa = 4.27KK398 pKa = 11.57NKK400 pKa = 7.1VTPMMWDD407 pKa = 2.87SGLCVGAGCLMCGWSVLVEE426 pKa = 3.86KK427 pKa = 10.69ASRR430 pKa = 11.84RR431 pKa = 11.84RR432 pKa = 11.84EE433 pKa = 3.7LSLFVAPRR441 pKa = 11.84AAATLLPRR449 pKa = 11.84VYY451 pKa = 10.58DD452 pKa = 3.03KK453 pKa = 11.04QYY455 pKa = 9.13QHH457 pKa = 7.02RR458 pKa = 11.84EE459 pKa = 3.72RR460 pKa = 11.84IAFAVCAAILSTCLQEE476 pKa = 4.21RR477 pKa = 11.84PEE479 pKa = 4.0MVRR482 pKa = 11.84GFFRR486 pKa = 11.84KK487 pKa = 9.73IGARR491 pKa = 11.84LIAA494 pKa = 4.7
Molecular weight: 56.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
4068950 |
51 |
8221 |
503.6 |
55.86 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
8.212 ± 0.024 | 1.155 ± 0.01 |
5.727 ± 0.017 | 6.347 ± 0.025 |
3.728 ± 0.016 | 6.686 ± 0.023 |
2.393 ± 0.01 | 4.982 ± 0.018 |
4.812 ± 0.023 | 9.035 ± 0.03 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.039 ± 0.009 | 3.711 ± 0.013 |
6.09 ± 0.03 | 3.964 ± 0.017 |
6.416 ± 0.024 | 8.631 ± 0.032 |
5.751 ± 0.018 | 6.175 ± 0.02 |
1.372 ± 0.009 | 2.773 ± 0.013 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
