
Capybara microvirus Cap3_SP_394
Taxonomy: Viruses; Monodnaviria; Sangervirae; Phixviricota; Malgrandaviricetes; Petitvirales; Microviridae; unclassified Microviridae
Average proteome isoelectric point is 5.46
Get precalculated fractions of proteins
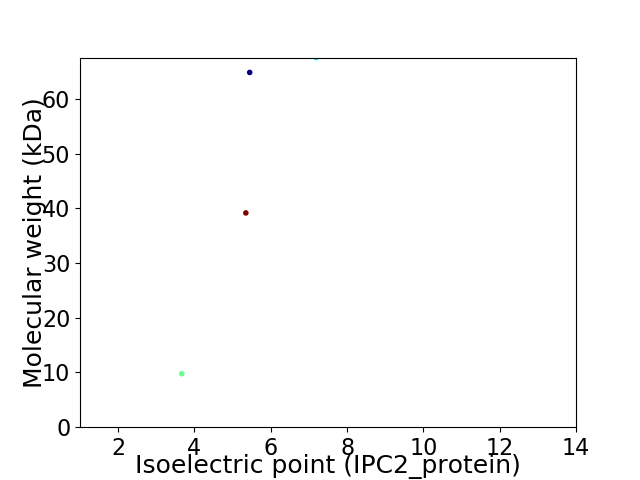
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A4P8W5B0|A0A4P8W5B0_9VIRU Uncharacterized protein OS=Capybara microvirus Cap3_SP_394 OX=2585444 PE=4 SV=1
MM1 pKa = 7.64VDD3 pKa = 3.65DD4 pKa = 5.44FLVEE8 pKa = 4.28SLPQEE13 pKa = 4.47DD14 pKa = 4.03GSYY17 pKa = 10.56ISVSVRR23 pKa = 11.84NQQKK27 pKa = 10.99DD28 pKa = 3.29YY29 pKa = 11.53GDD31 pKa = 3.83YY32 pKa = 11.43DD33 pKa = 3.94NYY35 pKa = 11.1TPDD38 pKa = 3.38ALSRR42 pKa = 11.84TGQTLPTMMMPDD54 pKa = 3.44GSSRR58 pKa = 11.84LEE60 pKa = 4.0ASSSLSSLLAQGEE73 pKa = 4.56QILSQPDD80 pKa = 3.71STPVTTDD87 pKa = 2.79NNGG90 pKa = 2.86
MM1 pKa = 7.64VDD3 pKa = 3.65DD4 pKa = 5.44FLVEE8 pKa = 4.28SLPQEE13 pKa = 4.47DD14 pKa = 4.03GSYY17 pKa = 10.56ISVSVRR23 pKa = 11.84NQQKK27 pKa = 10.99DD28 pKa = 3.29YY29 pKa = 11.53GDD31 pKa = 3.83YY32 pKa = 11.43DD33 pKa = 3.94NYY35 pKa = 11.1TPDD38 pKa = 3.38ALSRR42 pKa = 11.84TGQTLPTMMMPDD54 pKa = 3.44GSSRR58 pKa = 11.84LEE60 pKa = 4.0ASSSLSSLLAQGEE73 pKa = 4.56QILSQPDD80 pKa = 3.71STPVTTDD87 pKa = 2.79NNGG90 pKa = 2.86
Molecular weight: 9.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A4P8W4Q6|A0A4P8W4Q6_9VIRU Major capsid protein OS=Capybara microvirus Cap3_SP_394 OX=2585444 PE=3 SV=1
MM1 pKa = 7.47YY2 pKa = 9.46STIHH6 pKa = 6.23LKK8 pKa = 10.95SNAKK12 pKa = 8.78SWRR15 pKa = 11.84PLADD19 pKa = 2.93AAYY22 pKa = 9.71YY23 pKa = 9.87DD24 pKa = 4.39LPRR27 pKa = 11.84PTTFASKK34 pKa = 8.28TAARR38 pKa = 11.84YY39 pKa = 9.79GYY41 pKa = 9.22ICRR44 pKa = 11.84IKK46 pKa = 10.85AQLQDD51 pKa = 2.99TWRR54 pKa = 11.84SGGKK58 pKa = 10.07AFFYY62 pKa = 10.59TLTHH66 pKa = 6.26AQHH69 pKa = 6.52SLPHH73 pKa = 6.3FYY75 pKa = 10.79GIPVHH80 pKa = 7.08DD81 pKa = 3.55YY82 pKa = 10.49RR83 pKa = 11.84VINEE87 pKa = 4.3FFNCSGFSKK96 pKa = 10.84VLMRR100 pKa = 11.84RR101 pKa = 11.84HH102 pKa = 5.46NCRR105 pKa = 11.84IKK107 pKa = 10.35YY108 pKa = 9.67FLVSEE113 pKa = 4.71LGNGGEE119 pKa = 4.07SHH121 pKa = 6.08QYY123 pKa = 9.85KK124 pKa = 10.23GVRR127 pKa = 11.84GLLNNPHH134 pKa = 5.6YY135 pKa = 10.68HH136 pKa = 6.26VIFFLTSLDD145 pKa = 3.56DD146 pKa = 4.07HH147 pKa = 7.05LSVDD151 pKa = 3.33ITADD155 pKa = 3.26QLKK158 pKa = 10.36YY159 pKa = 10.4CVQKK163 pKa = 10.57YY164 pKa = 6.97WSGLPDD170 pKa = 3.51YY171 pKa = 11.11GVDD174 pKa = 3.46PKK176 pKa = 11.1VDD178 pKa = 4.39RR179 pKa = 11.84IRR181 pKa = 11.84DD182 pKa = 3.51NKK184 pKa = 10.11AALGAWIKK192 pKa = 10.85QYY194 pKa = 11.14GKK196 pKa = 10.94YY197 pKa = 10.02GLAWPGSLHH206 pKa = 6.34NGEE209 pKa = 5.27ILNQPDD215 pKa = 3.21SDD217 pKa = 3.89FANKK221 pKa = 9.27YY222 pKa = 6.82VCKK225 pKa = 10.53YY226 pKa = 8.6VVKK229 pKa = 10.71NHH231 pKa = 5.47ATYY234 pKa = 8.82TQAIRR239 pKa = 11.84LRR241 pKa = 11.84SEE243 pKa = 4.12LNRR246 pKa = 11.84LVYY249 pKa = 10.35GRR251 pKa = 11.84YY252 pKa = 9.7DD253 pKa = 3.37KK254 pKa = 11.41EE255 pKa = 3.95GVYY258 pKa = 9.95IDD260 pKa = 4.04GLLEE264 pKa = 4.55KK265 pKa = 10.1MMHH268 pKa = 6.18NPWQAYY274 pKa = 10.1AFMHH278 pKa = 6.74RR279 pKa = 11.84PKK281 pKa = 10.38KK282 pKa = 8.07WQCDD286 pKa = 3.65VYY288 pKa = 11.09RR289 pKa = 11.84DD290 pKa = 3.97YY291 pKa = 12.03NDD293 pKa = 3.22GSEE296 pKa = 4.12VDD298 pKa = 3.12KK299 pKa = 11.43SGYY302 pKa = 8.3TDD304 pKa = 4.4QEE306 pKa = 4.48ACLSWCLYY314 pKa = 9.54RR315 pKa = 11.84LKK317 pKa = 10.86SEE319 pKa = 4.13WQRR322 pKa = 11.84HH323 pKa = 4.24CTEE326 pKa = 4.49HH327 pKa = 6.43YY328 pKa = 9.43EE329 pKa = 4.15KK330 pKa = 10.87FIVRR334 pKa = 11.84QLALFRR340 pKa = 11.84SRR342 pKa = 11.84FAPHH346 pKa = 5.83IRR348 pKa = 11.84ASHH351 pKa = 5.6GVGVSALLEE360 pKa = 4.23VDD362 pKa = 5.63DD363 pKa = 6.53DD364 pKa = 4.91DD365 pKa = 4.18LTIDD369 pKa = 3.37VRR371 pKa = 11.84EE372 pKa = 4.09NGVVTKK378 pKa = 10.6HH379 pKa = 6.55RR380 pKa = 11.84ITGYY384 pKa = 9.79LARR387 pKa = 11.84KK388 pKa = 8.96KK389 pKa = 10.76LYY391 pKa = 10.46DD392 pKa = 3.61VVEE395 pKa = 4.41APGNQTRR402 pKa = 11.84YY403 pKa = 9.3VRR405 pKa = 11.84NSWYY409 pKa = 10.12KK410 pKa = 9.97QWLKK414 pKa = 11.31DD415 pKa = 3.53HH416 pKa = 6.6LAEE419 pKa = 6.11KK420 pKa = 9.9IDD422 pKa = 3.37KK423 pKa = 10.5CEE425 pKa = 3.84RR426 pKa = 11.84RR427 pKa = 11.84LRR429 pKa = 11.84DD430 pKa = 3.03LWYY433 pKa = 8.75NTDD436 pKa = 3.41INLLSKK442 pKa = 9.69ICEE445 pKa = 4.09YY446 pKa = 11.33NEE448 pKa = 3.49VDD450 pKa = 3.73VIPRR454 pKa = 11.84VVSDD458 pKa = 3.91FRR460 pKa = 11.84DD461 pKa = 3.36LAIYY465 pKa = 10.04ACVYY469 pKa = 9.98RR470 pKa = 11.84YY471 pKa = 9.82RR472 pKa = 11.84HH473 pKa = 5.63FHH475 pKa = 6.89RR476 pKa = 11.84DD477 pKa = 3.41DD478 pKa = 3.95PYY480 pKa = 11.61EE481 pKa = 3.98LYY483 pKa = 10.54NYY485 pKa = 9.63KK486 pKa = 10.12KK487 pKa = 10.73YY488 pKa = 11.05YY489 pKa = 7.57EE490 pKa = 4.09QCIDD494 pKa = 4.64APLEE498 pKa = 4.22DD499 pKa = 4.34TGCSYY504 pKa = 11.5AVYY507 pKa = 10.6QRR509 pKa = 11.84MGSFGQEE516 pKa = 3.62GTDD519 pKa = 3.48YY520 pKa = 11.13IPTNFVWPHH529 pKa = 5.59MEE531 pKa = 4.18ADD533 pKa = 3.78MEE535 pKa = 4.35VLDD538 pKa = 5.19MISDD542 pKa = 3.8YY543 pKa = 11.44LFAQIDD549 pKa = 3.64INRR552 pKa = 11.84DD553 pKa = 3.46KK554 pKa = 10.85EE555 pKa = 4.31WEE557 pKa = 4.24LKK559 pKa = 9.08SHH561 pKa = 6.09QMYY564 pKa = 10.23NQQQLAYY571 pKa = 9.92DD572 pKa = 3.87
MM1 pKa = 7.47YY2 pKa = 9.46STIHH6 pKa = 6.23LKK8 pKa = 10.95SNAKK12 pKa = 8.78SWRR15 pKa = 11.84PLADD19 pKa = 2.93AAYY22 pKa = 9.71YY23 pKa = 9.87DD24 pKa = 4.39LPRR27 pKa = 11.84PTTFASKK34 pKa = 8.28TAARR38 pKa = 11.84YY39 pKa = 9.79GYY41 pKa = 9.22ICRR44 pKa = 11.84IKK46 pKa = 10.85AQLQDD51 pKa = 2.99TWRR54 pKa = 11.84SGGKK58 pKa = 10.07AFFYY62 pKa = 10.59TLTHH66 pKa = 6.26AQHH69 pKa = 6.52SLPHH73 pKa = 6.3FYY75 pKa = 10.79GIPVHH80 pKa = 7.08DD81 pKa = 3.55YY82 pKa = 10.49RR83 pKa = 11.84VINEE87 pKa = 4.3FFNCSGFSKK96 pKa = 10.84VLMRR100 pKa = 11.84RR101 pKa = 11.84HH102 pKa = 5.46NCRR105 pKa = 11.84IKK107 pKa = 10.35YY108 pKa = 9.67FLVSEE113 pKa = 4.71LGNGGEE119 pKa = 4.07SHH121 pKa = 6.08QYY123 pKa = 9.85KK124 pKa = 10.23GVRR127 pKa = 11.84GLLNNPHH134 pKa = 5.6YY135 pKa = 10.68HH136 pKa = 6.26VIFFLTSLDD145 pKa = 3.56DD146 pKa = 4.07HH147 pKa = 7.05LSVDD151 pKa = 3.33ITADD155 pKa = 3.26QLKK158 pKa = 10.36YY159 pKa = 10.4CVQKK163 pKa = 10.57YY164 pKa = 6.97WSGLPDD170 pKa = 3.51YY171 pKa = 11.11GVDD174 pKa = 3.46PKK176 pKa = 11.1VDD178 pKa = 4.39RR179 pKa = 11.84IRR181 pKa = 11.84DD182 pKa = 3.51NKK184 pKa = 10.11AALGAWIKK192 pKa = 10.85QYY194 pKa = 11.14GKK196 pKa = 10.94YY197 pKa = 10.02GLAWPGSLHH206 pKa = 6.34NGEE209 pKa = 5.27ILNQPDD215 pKa = 3.21SDD217 pKa = 3.89FANKK221 pKa = 9.27YY222 pKa = 6.82VCKK225 pKa = 10.53YY226 pKa = 8.6VVKK229 pKa = 10.71NHH231 pKa = 5.47ATYY234 pKa = 8.82TQAIRR239 pKa = 11.84LRR241 pKa = 11.84SEE243 pKa = 4.12LNRR246 pKa = 11.84LVYY249 pKa = 10.35GRR251 pKa = 11.84YY252 pKa = 9.7DD253 pKa = 3.37KK254 pKa = 11.41EE255 pKa = 3.95GVYY258 pKa = 9.95IDD260 pKa = 4.04GLLEE264 pKa = 4.55KK265 pKa = 10.1MMHH268 pKa = 6.18NPWQAYY274 pKa = 10.1AFMHH278 pKa = 6.74RR279 pKa = 11.84PKK281 pKa = 10.38KK282 pKa = 8.07WQCDD286 pKa = 3.65VYY288 pKa = 11.09RR289 pKa = 11.84DD290 pKa = 3.97YY291 pKa = 12.03NDD293 pKa = 3.22GSEE296 pKa = 4.12VDD298 pKa = 3.12KK299 pKa = 11.43SGYY302 pKa = 8.3TDD304 pKa = 4.4QEE306 pKa = 4.48ACLSWCLYY314 pKa = 9.54RR315 pKa = 11.84LKK317 pKa = 10.86SEE319 pKa = 4.13WQRR322 pKa = 11.84HH323 pKa = 4.24CTEE326 pKa = 4.49HH327 pKa = 6.43YY328 pKa = 9.43EE329 pKa = 4.15KK330 pKa = 10.87FIVRR334 pKa = 11.84QLALFRR340 pKa = 11.84SRR342 pKa = 11.84FAPHH346 pKa = 5.83IRR348 pKa = 11.84ASHH351 pKa = 5.6GVGVSALLEE360 pKa = 4.23VDD362 pKa = 5.63DD363 pKa = 6.53DD364 pKa = 4.91DD365 pKa = 4.18LTIDD369 pKa = 3.37VRR371 pKa = 11.84EE372 pKa = 4.09NGVVTKK378 pKa = 10.6HH379 pKa = 6.55RR380 pKa = 11.84ITGYY384 pKa = 9.79LARR387 pKa = 11.84KK388 pKa = 8.96KK389 pKa = 10.76LYY391 pKa = 10.46DD392 pKa = 3.61VVEE395 pKa = 4.41APGNQTRR402 pKa = 11.84YY403 pKa = 9.3VRR405 pKa = 11.84NSWYY409 pKa = 10.12KK410 pKa = 9.97QWLKK414 pKa = 11.31DD415 pKa = 3.53HH416 pKa = 6.6LAEE419 pKa = 6.11KK420 pKa = 9.9IDD422 pKa = 3.37KK423 pKa = 10.5CEE425 pKa = 3.84RR426 pKa = 11.84RR427 pKa = 11.84LRR429 pKa = 11.84DD430 pKa = 3.03LWYY433 pKa = 8.75NTDD436 pKa = 3.41INLLSKK442 pKa = 9.69ICEE445 pKa = 4.09YY446 pKa = 11.33NEE448 pKa = 3.49VDD450 pKa = 3.73VIPRR454 pKa = 11.84VVSDD458 pKa = 3.91FRR460 pKa = 11.84DD461 pKa = 3.36LAIYY465 pKa = 10.04ACVYY469 pKa = 9.98RR470 pKa = 11.84YY471 pKa = 9.82RR472 pKa = 11.84HH473 pKa = 5.63FHH475 pKa = 6.89RR476 pKa = 11.84DD477 pKa = 3.41DD478 pKa = 3.95PYY480 pKa = 11.61EE481 pKa = 3.98LYY483 pKa = 10.54NYY485 pKa = 9.63KK486 pKa = 10.12KK487 pKa = 10.73YY488 pKa = 11.05YY489 pKa = 7.57EE490 pKa = 4.09QCIDD494 pKa = 4.64APLEE498 pKa = 4.22DD499 pKa = 4.34TGCSYY504 pKa = 11.5AVYY507 pKa = 10.6QRR509 pKa = 11.84MGSFGQEE516 pKa = 3.62GTDD519 pKa = 3.48YY520 pKa = 11.13IPTNFVWPHH529 pKa = 5.59MEE531 pKa = 4.18ADD533 pKa = 3.78MEE535 pKa = 4.35VLDD538 pKa = 5.19MISDD542 pKa = 3.8YY543 pKa = 11.44LFAQIDD549 pKa = 3.64INRR552 pKa = 11.84DD553 pKa = 3.46KK554 pKa = 10.85EE555 pKa = 4.31WEE557 pKa = 4.24LKK559 pKa = 9.08SHH561 pKa = 6.09QMYY564 pKa = 10.23NQQQLAYY571 pKa = 9.92DD572 pKa = 3.87
Molecular weight: 67.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1600 |
90 |
575 |
400.0 |
45.32 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.375 ± 0.967 | 1.188 ± 0.576 |
7.313 ± 0.686 | 3.938 ± 0.781 |
3.25 ± 0.855 | 6.563 ± 0.573 |
2.75 ± 0.724 | 5.625 ± 0.657 |
4.5 ± 0.933 | 8.438 ± 0.377 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.375 ± 0.42 | 5.063 ± 0.33 |
3.375 ± 0.456 | 6.188 ± 0.922 |
5.25 ± 0.739 | 8.0 ± 1.593 |
5.75 ± 0.954 | 5.438 ± 0.322 |
1.75 ± 0.378 | 5.875 ± 1.207 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
