
Pseudoalteromonas ruthenica
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Gammaproteobacteria; Alteromonadales; Pseudoalteromonadaceae; Pseudoalteromonas
Average proteome isoelectric point is 6.14
Get precalculated fractions of proteins
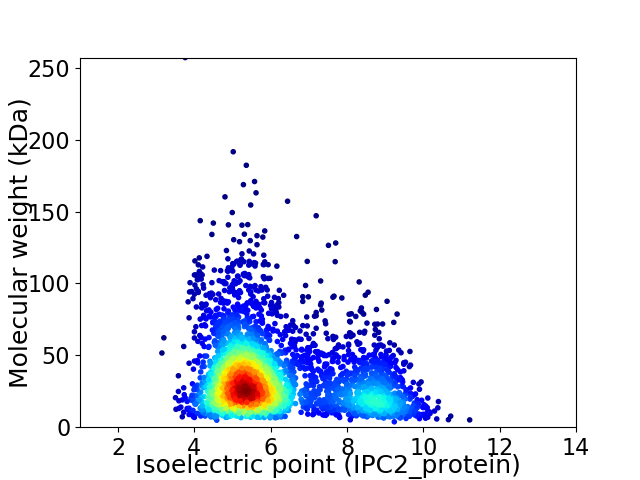
Virtual 2D-PAGE plot for 3470 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
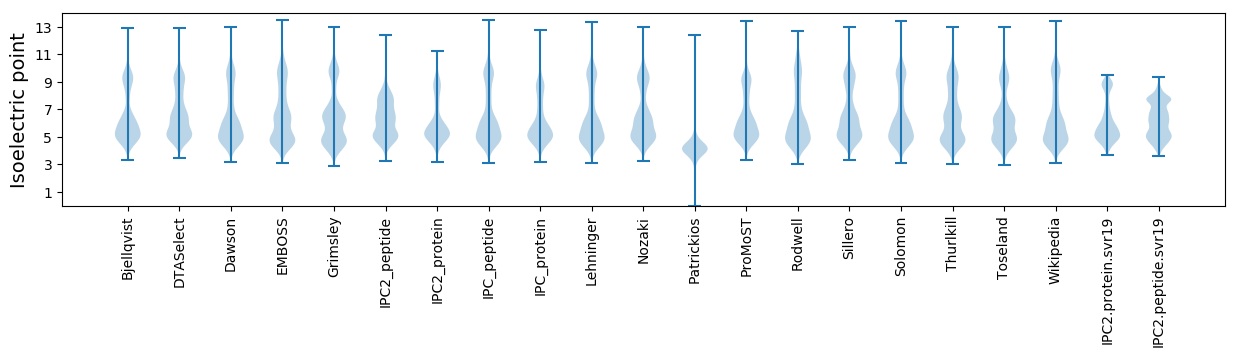
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0F4Q0M3|A0A0F4Q0M3_9GAMM Bifunctional protein FolD OS=Pseudoalteromonas ruthenica OX=151081 GN=folD PE=3 SV=1
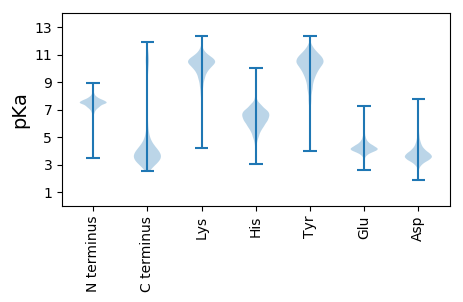
MM1 pKa = 7.65KK2 pKa = 9.01KK3 pKa = 8.22TLLGLSIAAALVSLPSFAEE22 pKa = 3.79VRR24 pKa = 11.84INGFASIVGGQTLDD38 pKa = 5.16DD39 pKa = 5.62DD40 pKa = 4.28DD41 pKa = 4.21TLYY44 pKa = 11.16GYY46 pKa = 11.07DD47 pKa = 3.59NDD49 pKa = 4.64FNMEE53 pKa = 4.01QEE55 pKa = 4.61SKK57 pKa = 10.78FALQVSADD65 pKa = 3.63LQEE68 pKa = 4.61RR69 pKa = 11.84LSATAQVIARR79 pKa = 11.84GEE81 pKa = 4.0NDD83 pKa = 2.83FDD85 pKa = 6.18AEE87 pKa = 4.27FEE89 pKa = 4.36WAYY92 pKa = 9.39LTYY95 pKa = 10.23EE96 pKa = 4.07ISDD99 pKa = 3.69NSQLSAGKK107 pKa = 8.75MRR109 pKa = 11.84IPFYY113 pKa = 10.42RR114 pKa = 11.84YY115 pKa = 9.92SDD117 pKa = 3.85FLDD120 pKa = 2.91VGYY123 pKa = 10.55AYY125 pKa = 10.49RR126 pKa = 11.84WVRR129 pKa = 11.84PPKK132 pKa = 10.23SVYY135 pKa = 9.93NLNFSTYY142 pKa = 9.73EE143 pKa = 3.96GLSYY147 pKa = 10.64LYY149 pKa = 10.44NSNIGEE155 pKa = 4.24WDD157 pKa = 3.34SSIQLMYY164 pKa = 11.05GSVNEE169 pKa = 5.11DD170 pKa = 2.59IAAVTEE176 pKa = 4.13SDD178 pKa = 3.27NVEE181 pKa = 3.81INDD184 pKa = 3.47AFGFNWTLSKK194 pKa = 11.11DD195 pKa = 3.19WFTARR200 pKa = 11.84AAYY203 pKa = 9.35ILSEE207 pKa = 4.02VSIDD211 pKa = 3.69VSNSAQLSALFSGLSGYY228 pKa = 10.02GLNEE232 pKa = 3.58QRR234 pKa = 11.84QDD236 pKa = 3.53LAVEE240 pKa = 4.15EE241 pKa = 4.7DD242 pKa = 3.42DD243 pKa = 5.78SYY245 pKa = 11.75FAGIGFGIDD254 pKa = 3.29YY255 pKa = 11.2NNFLFDD261 pKa = 4.47AEE263 pKa = 4.32YY264 pKa = 8.75TQYY267 pKa = 10.88EE268 pKa = 4.27IEE270 pKa = 4.65DD271 pKa = 3.84SFLAKK276 pKa = 10.05QSQYY280 pKa = 9.69YY281 pKa = 10.44ASVGYY286 pKa = 10.28RR287 pKa = 11.84IDD289 pKa = 3.29DD290 pKa = 3.65WTVHH294 pKa = 5.32FTYY297 pKa = 10.65EE298 pKa = 4.53NNDD301 pKa = 3.78DD302 pKa = 4.37EE303 pKa = 5.42NDD305 pKa = 3.63SSEE308 pKa = 4.93LNTIPAQIQTPNGPINVSTNPSDD331 pKa = 3.64PNAPLLRR338 pKa = 11.84DD339 pKa = 3.52VTNFALLSQRR349 pKa = 11.84AEE351 pKa = 4.24SNTYY355 pKa = 9.74TIGARR360 pKa = 11.84YY361 pKa = 9.43NFHH364 pKa = 7.22PSAAFKK370 pKa = 10.44IDD372 pKa = 3.22VSRR375 pKa = 11.84FEE377 pKa = 5.66DD378 pKa = 5.11DD379 pKa = 3.03ITNTEE384 pKa = 3.61VDD386 pKa = 4.41LVSMGVDD393 pKa = 3.41LVFF396 pKa = 5.28
MM1 pKa = 7.65KK2 pKa = 9.01KK3 pKa = 8.22TLLGLSIAAALVSLPSFAEE22 pKa = 3.79VRR24 pKa = 11.84INGFASIVGGQTLDD38 pKa = 5.16DD39 pKa = 5.62DD40 pKa = 4.28DD41 pKa = 4.21TLYY44 pKa = 11.16GYY46 pKa = 11.07DD47 pKa = 3.59NDD49 pKa = 4.64FNMEE53 pKa = 4.01QEE55 pKa = 4.61SKK57 pKa = 10.78FALQVSADD65 pKa = 3.63LQEE68 pKa = 4.61RR69 pKa = 11.84LSATAQVIARR79 pKa = 11.84GEE81 pKa = 4.0NDD83 pKa = 2.83FDD85 pKa = 6.18AEE87 pKa = 4.27FEE89 pKa = 4.36WAYY92 pKa = 9.39LTYY95 pKa = 10.23EE96 pKa = 4.07ISDD99 pKa = 3.69NSQLSAGKK107 pKa = 8.75MRR109 pKa = 11.84IPFYY113 pKa = 10.42RR114 pKa = 11.84YY115 pKa = 9.92SDD117 pKa = 3.85FLDD120 pKa = 2.91VGYY123 pKa = 10.55AYY125 pKa = 10.49RR126 pKa = 11.84WVRR129 pKa = 11.84PPKK132 pKa = 10.23SVYY135 pKa = 9.93NLNFSTYY142 pKa = 9.73EE143 pKa = 3.96GLSYY147 pKa = 10.64LYY149 pKa = 10.44NSNIGEE155 pKa = 4.24WDD157 pKa = 3.34SSIQLMYY164 pKa = 11.05GSVNEE169 pKa = 5.11DD170 pKa = 2.59IAAVTEE176 pKa = 4.13SDD178 pKa = 3.27NVEE181 pKa = 3.81INDD184 pKa = 3.47AFGFNWTLSKK194 pKa = 11.11DD195 pKa = 3.19WFTARR200 pKa = 11.84AAYY203 pKa = 9.35ILSEE207 pKa = 4.02VSIDD211 pKa = 3.69VSNSAQLSALFSGLSGYY228 pKa = 10.02GLNEE232 pKa = 3.58QRR234 pKa = 11.84QDD236 pKa = 3.53LAVEE240 pKa = 4.15EE241 pKa = 4.7DD242 pKa = 3.42DD243 pKa = 5.78SYY245 pKa = 11.75FAGIGFGIDD254 pKa = 3.29YY255 pKa = 11.2NNFLFDD261 pKa = 4.47AEE263 pKa = 4.32YY264 pKa = 8.75TQYY267 pKa = 10.88EE268 pKa = 4.27IEE270 pKa = 4.65DD271 pKa = 3.84SFLAKK276 pKa = 10.05QSQYY280 pKa = 9.69YY281 pKa = 10.44ASVGYY286 pKa = 10.28RR287 pKa = 11.84IDD289 pKa = 3.29DD290 pKa = 3.65WTVHH294 pKa = 5.32FTYY297 pKa = 10.65EE298 pKa = 4.53NNDD301 pKa = 3.78DD302 pKa = 4.37EE303 pKa = 5.42NDD305 pKa = 3.63SSEE308 pKa = 4.93LNTIPAQIQTPNGPINVSTNPSDD331 pKa = 3.64PNAPLLRR338 pKa = 11.84DD339 pKa = 3.52VTNFALLSQRR349 pKa = 11.84AEE351 pKa = 4.24SNTYY355 pKa = 9.74TIGARR360 pKa = 11.84YY361 pKa = 9.43NFHH364 pKa = 7.22PSAAFKK370 pKa = 10.44IDD372 pKa = 3.22VSRR375 pKa = 11.84FEE377 pKa = 5.66DD378 pKa = 5.11DD379 pKa = 3.03ITNTEE384 pKa = 3.61VDD386 pKa = 4.41LVSMGVDD393 pKa = 3.41LVFF396 pKa = 5.28
Molecular weight: 44.5 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0F4PYH9|A0A0F4PYH9_9GAMM Uncharacterized protein OS=Pseudoalteromonas ruthenica OX=151081 GN=TW72_08440 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
MM1 pKa = 7.45KK2 pKa = 9.51RR3 pKa = 11.84TFQPSVLKK11 pKa = 10.49RR12 pKa = 11.84KK13 pKa = 7.97RR14 pKa = 11.84THH16 pKa = 5.89GFRR19 pKa = 11.84ARR21 pKa = 11.84MASANGRR28 pKa = 11.84KK29 pKa = 9.04VLARR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84AKK37 pKa = 9.94GRR39 pKa = 11.84KK40 pKa = 8.67VLSAA44 pKa = 4.05
Molecular weight: 5.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1147547 |
34 |
2488 |
330.7 |
36.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.57 ± 0.044 | 1.013 ± 0.016 |
5.661 ± 0.035 | 6.034 ± 0.045 |
4.008 ± 0.028 | 6.689 ± 0.039 |
2.525 ± 0.028 | 5.682 ± 0.029 |
4.783 ± 0.042 | 10.505 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.405 ± 0.022 | 4.018 ± 0.027 |
3.838 ± 0.026 | 5.589 ± 0.049 |
4.764 ± 0.028 | 6.441 ± 0.035 |
5.095 ± 0.028 | 6.925 ± 0.037 |
1.213 ± 0.016 | 3.24 ± 0.027 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
