
Marivivens niveibacter
Taxonomy: cellular organisms; Bacteria; Proteobacteria; Alphaproteobacteria; Rhodobacterales; Rhodobacteraceae; Marivivens
Average proteome isoelectric point is 5.84
Get precalculated fractions of proteins
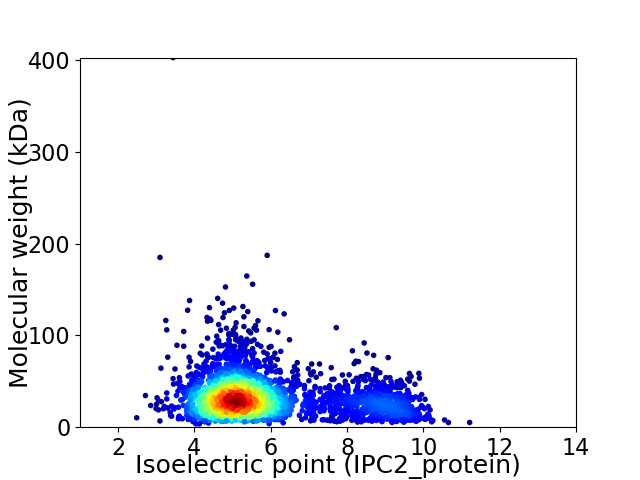
Virtual 2D-PAGE plot for 2990 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A251WZ41|A0A251WZ41_9RHOB 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase OS=Marivivens niveibacter OX=1930667 GN=hisA PE=3 SV=1
MM1 pKa = 7.17YY2 pKa = 7.79RR3 pKa = 11.84TLLATSAVLAISATASVAADD23 pKa = 3.36NAAVLDD29 pKa = 4.33TYY31 pKa = 11.95ANIAEE36 pKa = 4.46AGYY39 pKa = 11.13ADD41 pKa = 4.01SLATAKK47 pKa = 10.41VLQAAVNEE55 pKa = 4.41LVANPSEE62 pKa = 4.32DD63 pKa = 3.15TLAAARR69 pKa = 11.84SAWLDD74 pKa = 3.05ARR76 pKa = 11.84VPYY79 pKa = 10.14QQTEE83 pKa = 3.98VFRR86 pKa = 11.84FGNAIVDD93 pKa = 3.67DD94 pKa = 4.12WEE96 pKa = 4.58GLVNAWPLDD105 pKa = 3.65EE106 pKa = 5.54GLIDD110 pKa = 4.71YY111 pKa = 11.04VDD113 pKa = 3.49ASYY116 pKa = 11.35GGPTDD121 pKa = 3.64EE122 pKa = 6.08NEE124 pKa = 3.77FAALNVIANPTFTVAGVDD142 pKa = 3.07IDD144 pKa = 3.87ATNITPEE151 pKa = 5.03LIADD155 pKa = 4.21SLNEE159 pKa = 3.81ADD161 pKa = 5.08GIEE164 pKa = 4.37TNVARR169 pKa = 11.84GYY171 pKa = 10.46HH172 pKa = 6.42AIEE175 pKa = 4.05FLLWGQDD182 pKa = 4.15LNGTDD187 pKa = 3.88AGAGDD192 pKa = 4.54RR193 pKa = 11.84PYY195 pKa = 11.13TDD197 pKa = 3.85YY198 pKa = 11.96ADD200 pKa = 5.85GDD202 pKa = 3.98DD203 pKa = 4.35CTNGNCDD210 pKa = 3.02RR211 pKa = 11.84RR212 pKa = 11.84GDD214 pKa = 3.92YY215 pKa = 11.03LIAATDD221 pKa = 4.02LLVSDD226 pKa = 5.35LEE228 pKa = 4.75FITAAWAAEE237 pKa = 4.17GEE239 pKa = 4.25ARR241 pKa = 11.84AEE243 pKa = 3.92LTADD247 pKa = 3.44EE248 pKa = 4.4NAGIIAILTGMGSLSYY264 pKa = 11.07GEE266 pKa = 4.04QAGEE270 pKa = 3.95RR271 pKa = 11.84MRR273 pKa = 11.84LGLMLNDD280 pKa = 4.01PEE282 pKa = 4.81EE283 pKa = 4.14EE284 pKa = 4.02HH285 pKa = 7.9DD286 pKa = 3.95CFSDD290 pKa = 3.4NTHH293 pKa = 6.28NSHH296 pKa = 6.64YY297 pKa = 10.44YY298 pKa = 10.56DD299 pKa = 3.53GLGVQNVYY307 pKa = 10.27LGSYY311 pKa = 8.36TRR313 pKa = 11.84VDD315 pKa = 3.52GTVVSGPSLSEE326 pKa = 3.64LVAAANPEE334 pKa = 3.9LDD336 pKa = 3.98AEE338 pKa = 4.48LTAKK342 pKa = 10.85LNTTVAALGDD352 pKa = 3.52IVEE355 pKa = 4.3AAEE358 pKa = 4.21NGFSYY363 pKa = 11.68DD364 pKa = 3.26MMLEE368 pKa = 4.02RR369 pKa = 11.84GNAEE373 pKa = 3.65GEE375 pKa = 4.24ALVMGAVNGLIDD387 pKa = 3.51QTRR390 pKa = 11.84SIEE393 pKa = 4.23KK394 pKa = 10.02VVAALGLDD402 pKa = 4.24AIAFEE407 pKa = 5.46GSDD410 pKa = 4.15SLDD413 pKa = 3.48NPDD416 pKa = 5.36AVFQQ420 pKa = 4.04
MM1 pKa = 7.17YY2 pKa = 7.79RR3 pKa = 11.84TLLATSAVLAISATASVAADD23 pKa = 3.36NAAVLDD29 pKa = 4.33TYY31 pKa = 11.95ANIAEE36 pKa = 4.46AGYY39 pKa = 11.13ADD41 pKa = 4.01SLATAKK47 pKa = 10.41VLQAAVNEE55 pKa = 4.41LVANPSEE62 pKa = 4.32DD63 pKa = 3.15TLAAARR69 pKa = 11.84SAWLDD74 pKa = 3.05ARR76 pKa = 11.84VPYY79 pKa = 10.14QQTEE83 pKa = 3.98VFRR86 pKa = 11.84FGNAIVDD93 pKa = 3.67DD94 pKa = 4.12WEE96 pKa = 4.58GLVNAWPLDD105 pKa = 3.65EE106 pKa = 5.54GLIDD110 pKa = 4.71YY111 pKa = 11.04VDD113 pKa = 3.49ASYY116 pKa = 11.35GGPTDD121 pKa = 3.64EE122 pKa = 6.08NEE124 pKa = 3.77FAALNVIANPTFTVAGVDD142 pKa = 3.07IDD144 pKa = 3.87ATNITPEE151 pKa = 5.03LIADD155 pKa = 4.21SLNEE159 pKa = 3.81ADD161 pKa = 5.08GIEE164 pKa = 4.37TNVARR169 pKa = 11.84GYY171 pKa = 10.46HH172 pKa = 6.42AIEE175 pKa = 4.05FLLWGQDD182 pKa = 4.15LNGTDD187 pKa = 3.88AGAGDD192 pKa = 4.54RR193 pKa = 11.84PYY195 pKa = 11.13TDD197 pKa = 3.85YY198 pKa = 11.96ADD200 pKa = 5.85GDD202 pKa = 3.98DD203 pKa = 4.35CTNGNCDD210 pKa = 3.02RR211 pKa = 11.84RR212 pKa = 11.84GDD214 pKa = 3.92YY215 pKa = 11.03LIAATDD221 pKa = 4.02LLVSDD226 pKa = 5.35LEE228 pKa = 4.75FITAAWAAEE237 pKa = 4.17GEE239 pKa = 4.25ARR241 pKa = 11.84AEE243 pKa = 3.92LTADD247 pKa = 3.44EE248 pKa = 4.4NAGIIAILTGMGSLSYY264 pKa = 11.07GEE266 pKa = 4.04QAGEE270 pKa = 3.95RR271 pKa = 11.84MRR273 pKa = 11.84LGLMLNDD280 pKa = 4.01PEE282 pKa = 4.81EE283 pKa = 4.14EE284 pKa = 4.02HH285 pKa = 7.9DD286 pKa = 3.95CFSDD290 pKa = 3.4NTHH293 pKa = 6.28NSHH296 pKa = 6.64YY297 pKa = 10.44YY298 pKa = 10.56DD299 pKa = 3.53GLGVQNVYY307 pKa = 10.27LGSYY311 pKa = 8.36TRR313 pKa = 11.84VDD315 pKa = 3.52GTVVSGPSLSEE326 pKa = 3.64LVAAANPEE334 pKa = 3.9LDD336 pKa = 3.98AEE338 pKa = 4.48LTAKK342 pKa = 10.85LNTTVAALGDD352 pKa = 3.52IVEE355 pKa = 4.3AAEE358 pKa = 4.21NGFSYY363 pKa = 11.68DD364 pKa = 3.26MMLEE368 pKa = 4.02RR369 pKa = 11.84GNAEE373 pKa = 3.65GEE375 pKa = 4.24ALVMGAVNGLIDD387 pKa = 3.51QTRR390 pKa = 11.84SIEE393 pKa = 4.23KK394 pKa = 10.02VVAALGLDD402 pKa = 4.24AIAFEE407 pKa = 5.46GSDD410 pKa = 4.15SLDD413 pKa = 3.48NPDD416 pKa = 5.36AVFQQ420 pKa = 4.04
Molecular weight: 44.33 kDa
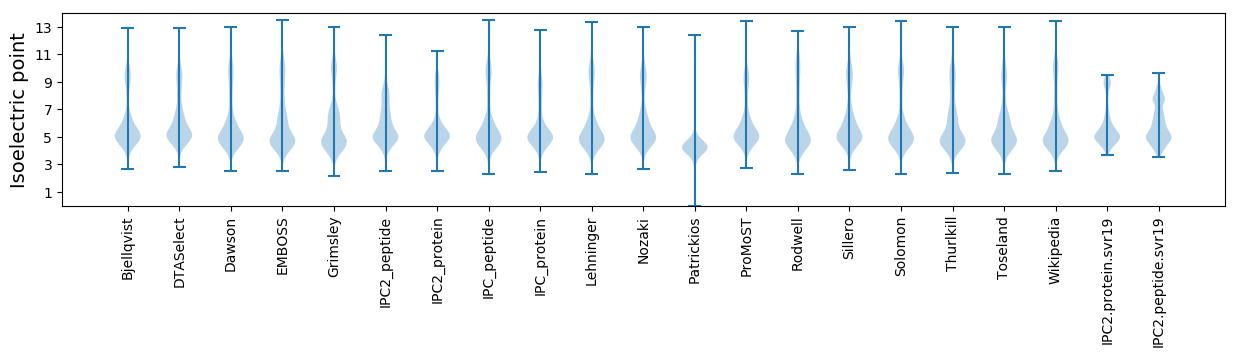
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A251WZ97|A0A251WZ97_9RHOB Uncharacterized protein OS=Marivivens niveibacter OX=1930667 GN=BVC71_11240 PE=4 SV=1
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.55ILNSRR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.31KK41 pKa = 10.65LSAA44 pKa = 3.91
MM1 pKa = 7.45KK2 pKa = 9.61RR3 pKa = 11.84TFQPSNRR10 pKa = 11.84VRR12 pKa = 11.84KK13 pKa = 8.99NRR15 pKa = 11.84HH16 pKa = 3.77GFRR19 pKa = 11.84ARR21 pKa = 11.84MATKK25 pKa = 10.37AGRR28 pKa = 11.84KK29 pKa = 8.55ILNSRR34 pKa = 11.84RR35 pKa = 11.84AQGRR39 pKa = 11.84KK40 pKa = 9.31KK41 pKa = 10.65LSAA44 pKa = 3.91
Molecular weight: 5.17 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
935407 |
32 |
3956 |
312.8 |
34.09 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.373 ± 0.058 | 0.891 ± 0.014 |
6.605 ± 0.044 | 5.797 ± 0.042 |
3.841 ± 0.029 | 8.22 ± 0.045 |
1.951 ± 0.023 | 5.993 ± 0.032 |
3.613 ± 0.04 | 9.288 ± 0.049 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.966 ± 0.025 | 3.22 ± 0.025 |
4.645 ± 0.029 | 3.349 ± 0.026 |
5.867 ± 0.046 | 5.432 ± 0.034 |
5.859 ± 0.051 | 7.368 ± 0.041 |
1.323 ± 0.017 | 2.4 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
