
Seal anellovirus 4
Taxonomy: Viruses; Anelloviridae; Nutorquevirus; Torque teno seal virus 4
Average proteome isoelectric point is 9.34
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0A7TXL6|A0A0A7TXL6_9VIRU Capsid protein OS=Seal anellovirus 4 OX=1566011 PE=3 SV=1
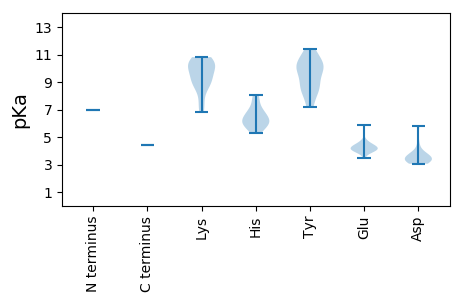
MM1 pKa = 7.0AWYY4 pKa = 8.39WRR6 pKa = 11.84WRR8 pKa = 11.84TRR10 pKa = 11.84RR11 pKa = 11.84WRR13 pKa = 11.84WRR15 pKa = 11.84PRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84FVRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84FYY28 pKa = 9.93PRR30 pKa = 11.84RR31 pKa = 11.84ANRR34 pKa = 11.84LRR36 pKa = 11.84YY37 pKa = 8.12NRR39 pKa = 11.84RR40 pKa = 11.84WVRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84FRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84HH54 pKa = 5.63APFTSKK60 pKa = 10.81VLLWNPSRR68 pKa = 11.84IVRR71 pKa = 11.84CKK73 pKa = 10.27IIGWHH78 pKa = 6.79PGLFCNQFNTIDD90 pKa = 3.48TCYY93 pKa = 11.14VNAIEE98 pKa = 5.85DD99 pKa = 3.76KK100 pKa = 10.08QTVQGFRR107 pKa = 11.84GGGVAFLALTLNMLYY122 pKa = 10.07RR123 pKa = 11.84EE124 pKa = 4.79HH125 pKa = 8.03IFGRR129 pKa = 11.84NFWTRR134 pKa = 11.84SNEE137 pKa = 3.99GFDD140 pKa = 3.34LAQYY144 pKa = 10.36RR145 pKa = 11.84GTRR148 pKa = 11.84VTFFPHH154 pKa = 5.27YY155 pKa = 8.25QLPYY159 pKa = 9.1IVTWLRR165 pKa = 11.84DD166 pKa = 3.13FATPVDD172 pKa = 3.55KK173 pKa = 10.69HH174 pKa = 6.85IQDD177 pKa = 3.58MHH179 pKa = 5.84PQRR182 pKa = 11.84ALFGPNHH189 pKa = 6.32KK190 pKa = 10.58VILPRR195 pKa = 11.84VFGRR199 pKa = 11.84KK200 pKa = 9.09RR201 pKa = 11.84KK202 pKa = 7.18WSLYY206 pKa = 9.51IRR208 pKa = 11.84PPPLNEE214 pKa = 3.9RR215 pKa = 11.84KK216 pKa = 8.87WYY218 pKa = 11.37SMVSWCNVVIAKK230 pKa = 10.21IGISFINLTSPLLHH244 pKa = 6.32QEE246 pKa = 4.34SPFPAVFCGYY256 pKa = 8.63TMQSGSFGEE265 pKa = 4.61PPRR268 pKa = 11.84CMLDD272 pKa = 3.3PEE274 pKa = 4.67GLPKK278 pKa = 10.25VRR280 pKa = 11.84EE281 pKa = 3.92NPTWHH286 pKa = 6.09VGNYY290 pKa = 9.08NLPCCYY296 pKa = 10.01RR297 pKa = 11.84PLWDD301 pKa = 5.79DD302 pKa = 3.35GTDD305 pKa = 3.21NYY307 pKa = 11.05VLMNYY312 pKa = 7.17TNKK315 pKa = 9.85PPSVQGTYY323 pKa = 10.32RR324 pKa = 11.84PTGTGQEE331 pKa = 4.05VLIWQAKK338 pKa = 7.79NRR340 pKa = 11.84HH341 pKa = 5.79ALLAMVLRR349 pKa = 11.84FNRR352 pKa = 11.84IKK354 pKa = 10.64RR355 pKa = 11.84NYY357 pKa = 8.46LCKK360 pKa = 10.12PLATWQIFWPSVMGMYY376 pKa = 9.74WYY378 pKa = 10.7VDD380 pKa = 3.31QARR383 pKa = 11.84VLEE386 pKa = 4.4GTVDD390 pKa = 3.95TIVPDD395 pKa = 3.27MSITVPEE402 pKa = 4.12EE403 pKa = 3.96LPTKK407 pKa = 8.63TNRR410 pKa = 11.84SWIIFTPPRR419 pKa = 11.84GEE421 pKa = 3.76NMAKK425 pKa = 9.56IRR427 pKa = 11.84EE428 pKa = 4.51HH429 pKa = 6.01IHH431 pKa = 6.16GNNIMTWPTLPQVKK445 pKa = 8.73TICSRR450 pKa = 11.84IAGSAGFTIGYY461 pKa = 10.16GEE463 pKa = 4.34MPIKK467 pKa = 9.08TANVPFFYY475 pKa = 10.29KK476 pKa = 10.48SYY478 pKa = 8.77WRR480 pKa = 11.84WGGSFADD487 pKa = 3.36ADD489 pKa = 3.81TRR491 pKa = 11.84VRR493 pKa = 11.84NPCVEE498 pKa = 4.25LPMATGTDD506 pKa = 3.09RR507 pKa = 11.84LGVQIRR513 pKa = 11.84DD514 pKa = 3.49PATVSLANIHH524 pKa = 6.48PWDD527 pKa = 4.53LEE529 pKa = 4.13NGSITKK535 pKa = 9.54PAMLRR540 pKa = 11.84ILSSILGPGSHH551 pKa = 7.04LPGAPTADD559 pKa = 3.56PQLPQAKK566 pKa = 9.32EE567 pKa = 4.12GEE569 pKa = 4.3EE570 pKa = 4.13LPEE573 pKa = 4.07QSSSSGDD580 pKa = 3.06SDD582 pKa = 3.78TPTEE586 pKa = 4.21EE587 pKa = 4.16EE588 pKa = 4.42PPTSEE593 pKa = 4.43EE594 pKa = 3.99EE595 pKa = 4.28QEE597 pKa = 4.35TSQVWRR603 pKa = 11.84RR604 pKa = 11.84KK605 pKa = 6.85LQRR608 pKa = 11.84MGKK611 pKa = 8.88RR612 pKa = 11.84LRR614 pKa = 11.84RR615 pKa = 11.84EE616 pKa = 3.47QQYY619 pKa = 10.18RR620 pKa = 11.84RR621 pKa = 11.84RR622 pKa = 11.84VKK624 pKa = 10.32QRR626 pKa = 11.84LLEE629 pKa = 4.03LSGYY633 pKa = 7.15QHH635 pKa = 7.68PSTQFMEE642 pKa = 4.41
MM1 pKa = 7.0AWYY4 pKa = 8.39WRR6 pKa = 11.84WRR8 pKa = 11.84TRR10 pKa = 11.84RR11 pKa = 11.84WRR13 pKa = 11.84WRR15 pKa = 11.84PRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84FVRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84FYY28 pKa = 9.93PRR30 pKa = 11.84RR31 pKa = 11.84ANRR34 pKa = 11.84LRR36 pKa = 11.84YY37 pKa = 8.12NRR39 pKa = 11.84RR40 pKa = 11.84WVRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84FRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84HH54 pKa = 5.63APFTSKK60 pKa = 10.81VLLWNPSRR68 pKa = 11.84IVRR71 pKa = 11.84CKK73 pKa = 10.27IIGWHH78 pKa = 6.79PGLFCNQFNTIDD90 pKa = 3.48TCYY93 pKa = 11.14VNAIEE98 pKa = 5.85DD99 pKa = 3.76KK100 pKa = 10.08QTVQGFRR107 pKa = 11.84GGGVAFLALTLNMLYY122 pKa = 10.07RR123 pKa = 11.84EE124 pKa = 4.79HH125 pKa = 8.03IFGRR129 pKa = 11.84NFWTRR134 pKa = 11.84SNEE137 pKa = 3.99GFDD140 pKa = 3.34LAQYY144 pKa = 10.36RR145 pKa = 11.84GTRR148 pKa = 11.84VTFFPHH154 pKa = 5.27YY155 pKa = 8.25QLPYY159 pKa = 9.1IVTWLRR165 pKa = 11.84DD166 pKa = 3.13FATPVDD172 pKa = 3.55KK173 pKa = 10.69HH174 pKa = 6.85IQDD177 pKa = 3.58MHH179 pKa = 5.84PQRR182 pKa = 11.84ALFGPNHH189 pKa = 6.32KK190 pKa = 10.58VILPRR195 pKa = 11.84VFGRR199 pKa = 11.84KK200 pKa = 9.09RR201 pKa = 11.84KK202 pKa = 7.18WSLYY206 pKa = 9.51IRR208 pKa = 11.84PPPLNEE214 pKa = 3.9RR215 pKa = 11.84KK216 pKa = 8.87WYY218 pKa = 11.37SMVSWCNVVIAKK230 pKa = 10.21IGISFINLTSPLLHH244 pKa = 6.32QEE246 pKa = 4.34SPFPAVFCGYY256 pKa = 8.63TMQSGSFGEE265 pKa = 4.61PPRR268 pKa = 11.84CMLDD272 pKa = 3.3PEE274 pKa = 4.67GLPKK278 pKa = 10.25VRR280 pKa = 11.84EE281 pKa = 3.92NPTWHH286 pKa = 6.09VGNYY290 pKa = 9.08NLPCCYY296 pKa = 10.01RR297 pKa = 11.84PLWDD301 pKa = 5.79DD302 pKa = 3.35GTDD305 pKa = 3.21NYY307 pKa = 11.05VLMNYY312 pKa = 7.17TNKK315 pKa = 9.85PPSVQGTYY323 pKa = 10.32RR324 pKa = 11.84PTGTGQEE331 pKa = 4.05VLIWQAKK338 pKa = 7.79NRR340 pKa = 11.84HH341 pKa = 5.79ALLAMVLRR349 pKa = 11.84FNRR352 pKa = 11.84IKK354 pKa = 10.64RR355 pKa = 11.84NYY357 pKa = 8.46LCKK360 pKa = 10.12PLATWQIFWPSVMGMYY376 pKa = 9.74WYY378 pKa = 10.7VDD380 pKa = 3.31QARR383 pKa = 11.84VLEE386 pKa = 4.4GTVDD390 pKa = 3.95TIVPDD395 pKa = 3.27MSITVPEE402 pKa = 4.12EE403 pKa = 3.96LPTKK407 pKa = 8.63TNRR410 pKa = 11.84SWIIFTPPRR419 pKa = 11.84GEE421 pKa = 3.76NMAKK425 pKa = 9.56IRR427 pKa = 11.84EE428 pKa = 4.51HH429 pKa = 6.01IHH431 pKa = 6.16GNNIMTWPTLPQVKK445 pKa = 8.73TICSRR450 pKa = 11.84IAGSAGFTIGYY461 pKa = 10.16GEE463 pKa = 4.34MPIKK467 pKa = 9.08TANVPFFYY475 pKa = 10.29KK476 pKa = 10.48SYY478 pKa = 8.77WRR480 pKa = 11.84WGGSFADD487 pKa = 3.36ADD489 pKa = 3.81TRR491 pKa = 11.84VRR493 pKa = 11.84NPCVEE498 pKa = 4.25LPMATGTDD506 pKa = 3.09RR507 pKa = 11.84LGVQIRR513 pKa = 11.84DD514 pKa = 3.49PATVSLANIHH524 pKa = 6.48PWDD527 pKa = 4.53LEE529 pKa = 4.13NGSITKK535 pKa = 9.54PAMLRR540 pKa = 11.84ILSSILGPGSHH551 pKa = 7.04LPGAPTADD559 pKa = 3.56PQLPQAKK566 pKa = 9.32EE567 pKa = 4.12GEE569 pKa = 4.3EE570 pKa = 4.13LPEE573 pKa = 4.07QSSSSGDD580 pKa = 3.06SDD582 pKa = 3.78TPTEE586 pKa = 4.21EE587 pKa = 4.16EE588 pKa = 4.42PPTSEE593 pKa = 4.43EE594 pKa = 3.99EE595 pKa = 4.28QEE597 pKa = 4.35TSQVWRR603 pKa = 11.84RR604 pKa = 11.84KK605 pKa = 6.85LQRR608 pKa = 11.84MGKK611 pKa = 8.88RR612 pKa = 11.84LRR614 pKa = 11.84RR615 pKa = 11.84EE616 pKa = 3.47QQYY619 pKa = 10.18RR620 pKa = 11.84RR621 pKa = 11.84RR622 pKa = 11.84VKK624 pKa = 10.32QRR626 pKa = 11.84LLEE629 pKa = 4.03LSGYY633 pKa = 7.15QHH635 pKa = 7.68PSTQFMEE642 pKa = 4.41
Molecular weight: 75.49 kDa
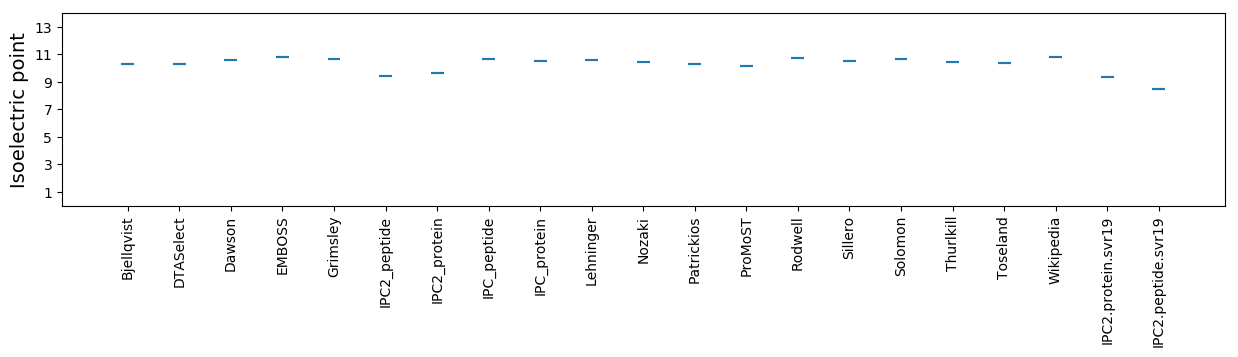
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0A7TXL6|A0A0A7TXL6_9VIRU Capsid protein OS=Seal anellovirus 4 OX=1566011 PE=3 SV=1
MM1 pKa = 7.0AWYY4 pKa = 8.39WRR6 pKa = 11.84WRR8 pKa = 11.84TRR10 pKa = 11.84RR11 pKa = 11.84WRR13 pKa = 11.84WRR15 pKa = 11.84PRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84FVRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84FYY28 pKa = 9.93PRR30 pKa = 11.84RR31 pKa = 11.84ANRR34 pKa = 11.84LRR36 pKa = 11.84YY37 pKa = 8.12NRR39 pKa = 11.84RR40 pKa = 11.84WVRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84FRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84HH54 pKa = 5.63APFTSKK60 pKa = 10.81VLLWNPSRR68 pKa = 11.84IVRR71 pKa = 11.84CKK73 pKa = 10.27IIGWHH78 pKa = 6.79PGLFCNQFNTIDD90 pKa = 3.48TCYY93 pKa = 11.14VNAIEE98 pKa = 5.85DD99 pKa = 3.76KK100 pKa = 10.08QTVQGFRR107 pKa = 11.84GGGVAFLALTLNMLYY122 pKa = 10.07RR123 pKa = 11.84EE124 pKa = 4.79HH125 pKa = 8.03IFGRR129 pKa = 11.84NFWTRR134 pKa = 11.84SNEE137 pKa = 3.99GFDD140 pKa = 3.34LAQYY144 pKa = 10.36RR145 pKa = 11.84GTRR148 pKa = 11.84VTFFPHH154 pKa = 5.27YY155 pKa = 8.25QLPYY159 pKa = 9.1IVTWLRR165 pKa = 11.84DD166 pKa = 3.13FATPVDD172 pKa = 3.55KK173 pKa = 10.69HH174 pKa = 6.85IQDD177 pKa = 3.58MHH179 pKa = 5.84PQRR182 pKa = 11.84ALFGPNHH189 pKa = 6.32KK190 pKa = 10.58VILPRR195 pKa = 11.84VFGRR199 pKa = 11.84KK200 pKa = 9.09RR201 pKa = 11.84KK202 pKa = 7.18WSLYY206 pKa = 9.51IRR208 pKa = 11.84PPPLNEE214 pKa = 3.9RR215 pKa = 11.84KK216 pKa = 8.87WYY218 pKa = 11.37SMVSWCNVVIAKK230 pKa = 10.21IGISFINLTSPLLHH244 pKa = 6.32QEE246 pKa = 4.34SPFPAVFCGYY256 pKa = 8.63TMQSGSFGEE265 pKa = 4.61PPRR268 pKa = 11.84CMLDD272 pKa = 3.3PEE274 pKa = 4.67GLPKK278 pKa = 10.25VRR280 pKa = 11.84EE281 pKa = 3.92NPTWHH286 pKa = 6.09VGNYY290 pKa = 9.08NLPCCYY296 pKa = 10.01RR297 pKa = 11.84PLWDD301 pKa = 5.79DD302 pKa = 3.35GTDD305 pKa = 3.21NYY307 pKa = 11.05VLMNYY312 pKa = 7.17TNKK315 pKa = 9.85PPSVQGTYY323 pKa = 10.32RR324 pKa = 11.84PTGTGQEE331 pKa = 4.05VLIWQAKK338 pKa = 7.79NRR340 pKa = 11.84HH341 pKa = 5.79ALLAMVLRR349 pKa = 11.84FNRR352 pKa = 11.84IKK354 pKa = 10.64RR355 pKa = 11.84NYY357 pKa = 8.46LCKK360 pKa = 10.12PLATWQIFWPSVMGMYY376 pKa = 9.74WYY378 pKa = 10.7VDD380 pKa = 3.31QARR383 pKa = 11.84VLEE386 pKa = 4.4GTVDD390 pKa = 3.95TIVPDD395 pKa = 3.27MSITVPEE402 pKa = 4.12EE403 pKa = 3.96LPTKK407 pKa = 8.63TNRR410 pKa = 11.84SWIIFTPPRR419 pKa = 11.84GEE421 pKa = 3.76NMAKK425 pKa = 9.56IRR427 pKa = 11.84EE428 pKa = 4.51HH429 pKa = 6.01IHH431 pKa = 6.16GNNIMTWPTLPQVKK445 pKa = 8.73TICSRR450 pKa = 11.84IAGSAGFTIGYY461 pKa = 10.16GEE463 pKa = 4.34MPIKK467 pKa = 9.08TANVPFFYY475 pKa = 10.29KK476 pKa = 10.48SYY478 pKa = 8.77WRR480 pKa = 11.84WGGSFADD487 pKa = 3.36ADD489 pKa = 3.81TRR491 pKa = 11.84VRR493 pKa = 11.84NPCVEE498 pKa = 4.25LPMATGTDD506 pKa = 3.09RR507 pKa = 11.84LGVQIRR513 pKa = 11.84DD514 pKa = 3.49PATVSLANIHH524 pKa = 6.48PWDD527 pKa = 4.53LEE529 pKa = 4.13NGSITKK535 pKa = 9.54PAMLRR540 pKa = 11.84ILSSILGPGSHH551 pKa = 7.04LPGAPTADD559 pKa = 3.56PQLPQAKK566 pKa = 9.32EE567 pKa = 4.12GEE569 pKa = 4.3EE570 pKa = 4.13LPEE573 pKa = 4.07QSSSSGDD580 pKa = 3.06SDD582 pKa = 3.78TPTEE586 pKa = 4.21EE587 pKa = 4.16EE588 pKa = 4.42PPTSEE593 pKa = 4.43EE594 pKa = 3.99EE595 pKa = 4.28QEE597 pKa = 4.35TSQVWRR603 pKa = 11.84RR604 pKa = 11.84KK605 pKa = 6.85LQRR608 pKa = 11.84MGKK611 pKa = 8.88RR612 pKa = 11.84LRR614 pKa = 11.84RR615 pKa = 11.84EE616 pKa = 3.47QQYY619 pKa = 10.18RR620 pKa = 11.84RR621 pKa = 11.84RR622 pKa = 11.84VKK624 pKa = 10.32QRR626 pKa = 11.84LLEE629 pKa = 4.03LSGYY633 pKa = 7.15QHH635 pKa = 7.68PSTQFMEE642 pKa = 4.41
MM1 pKa = 7.0AWYY4 pKa = 8.39WRR6 pKa = 11.84WRR8 pKa = 11.84TRR10 pKa = 11.84RR11 pKa = 11.84WRR13 pKa = 11.84WRR15 pKa = 11.84PRR17 pKa = 11.84RR18 pKa = 11.84RR19 pKa = 11.84FVRR22 pKa = 11.84RR23 pKa = 11.84RR24 pKa = 11.84RR25 pKa = 11.84RR26 pKa = 11.84FYY28 pKa = 9.93PRR30 pKa = 11.84RR31 pKa = 11.84ANRR34 pKa = 11.84LRR36 pKa = 11.84YY37 pKa = 8.12NRR39 pKa = 11.84RR40 pKa = 11.84WVRR43 pKa = 11.84RR44 pKa = 11.84RR45 pKa = 11.84RR46 pKa = 11.84FRR48 pKa = 11.84RR49 pKa = 11.84RR50 pKa = 11.84RR51 pKa = 11.84RR52 pKa = 11.84RR53 pKa = 11.84HH54 pKa = 5.63APFTSKK60 pKa = 10.81VLLWNPSRR68 pKa = 11.84IVRR71 pKa = 11.84CKK73 pKa = 10.27IIGWHH78 pKa = 6.79PGLFCNQFNTIDD90 pKa = 3.48TCYY93 pKa = 11.14VNAIEE98 pKa = 5.85DD99 pKa = 3.76KK100 pKa = 10.08QTVQGFRR107 pKa = 11.84GGGVAFLALTLNMLYY122 pKa = 10.07RR123 pKa = 11.84EE124 pKa = 4.79HH125 pKa = 8.03IFGRR129 pKa = 11.84NFWTRR134 pKa = 11.84SNEE137 pKa = 3.99GFDD140 pKa = 3.34LAQYY144 pKa = 10.36RR145 pKa = 11.84GTRR148 pKa = 11.84VTFFPHH154 pKa = 5.27YY155 pKa = 8.25QLPYY159 pKa = 9.1IVTWLRR165 pKa = 11.84DD166 pKa = 3.13FATPVDD172 pKa = 3.55KK173 pKa = 10.69HH174 pKa = 6.85IQDD177 pKa = 3.58MHH179 pKa = 5.84PQRR182 pKa = 11.84ALFGPNHH189 pKa = 6.32KK190 pKa = 10.58VILPRR195 pKa = 11.84VFGRR199 pKa = 11.84KK200 pKa = 9.09RR201 pKa = 11.84KK202 pKa = 7.18WSLYY206 pKa = 9.51IRR208 pKa = 11.84PPPLNEE214 pKa = 3.9RR215 pKa = 11.84KK216 pKa = 8.87WYY218 pKa = 11.37SMVSWCNVVIAKK230 pKa = 10.21IGISFINLTSPLLHH244 pKa = 6.32QEE246 pKa = 4.34SPFPAVFCGYY256 pKa = 8.63TMQSGSFGEE265 pKa = 4.61PPRR268 pKa = 11.84CMLDD272 pKa = 3.3PEE274 pKa = 4.67GLPKK278 pKa = 10.25VRR280 pKa = 11.84EE281 pKa = 3.92NPTWHH286 pKa = 6.09VGNYY290 pKa = 9.08NLPCCYY296 pKa = 10.01RR297 pKa = 11.84PLWDD301 pKa = 5.79DD302 pKa = 3.35GTDD305 pKa = 3.21NYY307 pKa = 11.05VLMNYY312 pKa = 7.17TNKK315 pKa = 9.85PPSVQGTYY323 pKa = 10.32RR324 pKa = 11.84PTGTGQEE331 pKa = 4.05VLIWQAKK338 pKa = 7.79NRR340 pKa = 11.84HH341 pKa = 5.79ALLAMVLRR349 pKa = 11.84FNRR352 pKa = 11.84IKK354 pKa = 10.64RR355 pKa = 11.84NYY357 pKa = 8.46LCKK360 pKa = 10.12PLATWQIFWPSVMGMYY376 pKa = 9.74WYY378 pKa = 10.7VDD380 pKa = 3.31QARR383 pKa = 11.84VLEE386 pKa = 4.4GTVDD390 pKa = 3.95TIVPDD395 pKa = 3.27MSITVPEE402 pKa = 4.12EE403 pKa = 3.96LPTKK407 pKa = 8.63TNRR410 pKa = 11.84SWIIFTPPRR419 pKa = 11.84GEE421 pKa = 3.76NMAKK425 pKa = 9.56IRR427 pKa = 11.84EE428 pKa = 4.51HH429 pKa = 6.01IHH431 pKa = 6.16GNNIMTWPTLPQVKK445 pKa = 8.73TICSRR450 pKa = 11.84IAGSAGFTIGYY461 pKa = 10.16GEE463 pKa = 4.34MPIKK467 pKa = 9.08TANVPFFYY475 pKa = 10.29KK476 pKa = 10.48SYY478 pKa = 8.77WRR480 pKa = 11.84WGGSFADD487 pKa = 3.36ADD489 pKa = 3.81TRR491 pKa = 11.84VRR493 pKa = 11.84NPCVEE498 pKa = 4.25LPMATGTDD506 pKa = 3.09RR507 pKa = 11.84LGVQIRR513 pKa = 11.84DD514 pKa = 3.49PATVSLANIHH524 pKa = 6.48PWDD527 pKa = 4.53LEE529 pKa = 4.13NGSITKK535 pKa = 9.54PAMLRR540 pKa = 11.84ILSSILGPGSHH551 pKa = 7.04LPGAPTADD559 pKa = 3.56PQLPQAKK566 pKa = 9.32EE567 pKa = 4.12GEE569 pKa = 4.3EE570 pKa = 4.13LPEE573 pKa = 4.07QSSSSGDD580 pKa = 3.06SDD582 pKa = 3.78TPTEE586 pKa = 4.21EE587 pKa = 4.16EE588 pKa = 4.42PPTSEE593 pKa = 4.43EE594 pKa = 3.99EE595 pKa = 4.28QEE597 pKa = 4.35TSQVWRR603 pKa = 11.84RR604 pKa = 11.84KK605 pKa = 6.85LQRR608 pKa = 11.84MGKK611 pKa = 8.88RR612 pKa = 11.84LRR614 pKa = 11.84RR615 pKa = 11.84EE616 pKa = 3.47QQYY619 pKa = 10.18RR620 pKa = 11.84RR621 pKa = 11.84RR622 pKa = 11.84VKK624 pKa = 10.32QRR626 pKa = 11.84LLEE629 pKa = 4.03LSGYY633 pKa = 7.15QHH635 pKa = 7.68PSTQFMEE642 pKa = 4.41
Molecular weight: 75.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
642 |
642 |
642 |
642.0 |
75.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
4.517 ± 0.0 | 1.713 ± 0.0 |
3.271 ± 0.0 | 4.829 ± 0.0 |
4.361 ± 0.0 | 6.386 ± 0.0 |
2.336 ± 0.0 | 5.14 ± 0.0 |
3.738 ± 0.0 | 7.321 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.804 ± 0.0 | 4.673 ± 0.0 |
8.255 ± 0.0 | 4.206 ± 0.0 |
11.526 ± 0.0 | 5.14 ± 0.0 |
6.542 ± 0.0 | 5.607 ± 0.0 |
3.894 ± 0.0 | 3.738 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
