
Bat associated circovirus 1
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; Circovirus
Average proteome isoelectric point is 8.45
Get precalculated fractions of proteins
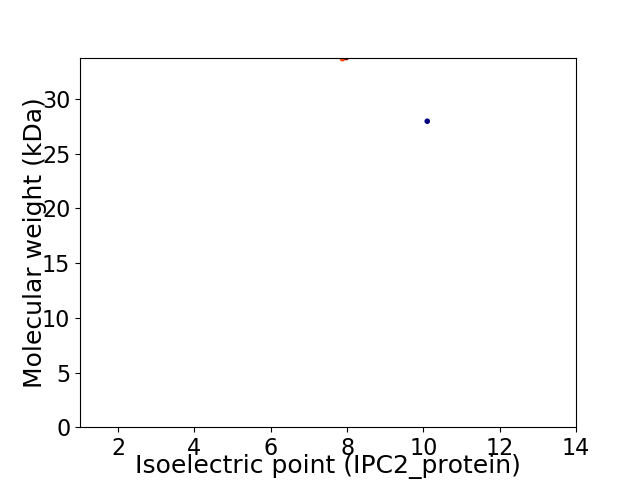
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
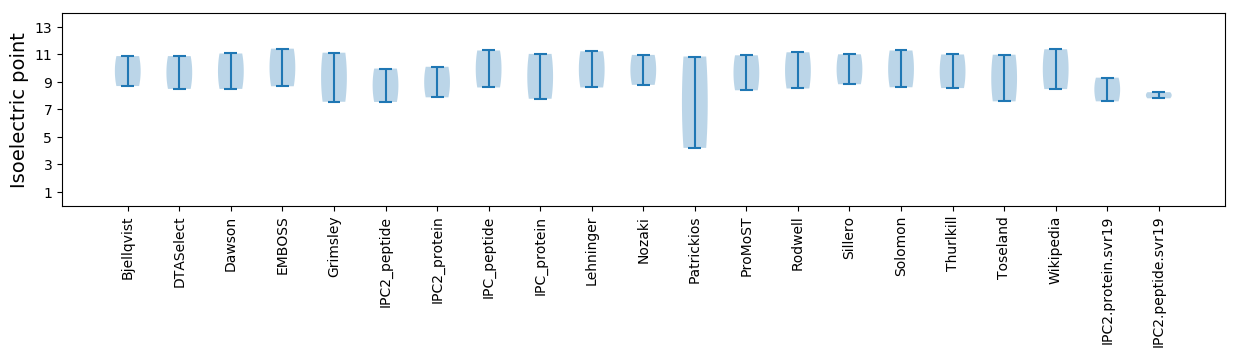
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|R4L9E4|R4L9E4_9CIRC Capsid protein OS=Bat associated circovirus 1 OX=1868218 GN=cap PE=3 SV=1
MM1 pKa = 7.46EE2 pKa = 4.57MAGHH6 pKa = 6.25PCEE9 pKa = 4.97RR10 pKa = 11.84YY11 pKa = 10.13CFTINNYY18 pKa = 9.0SEE20 pKa = 4.06EE21 pKa = 4.62DD22 pKa = 3.4IEE24 pKa = 4.23AVKK27 pKa = 10.96AFLVPDD33 pKa = 3.56NAEE36 pKa = 3.78YY37 pKa = 10.58AIVGKK42 pKa = 10.44EE43 pKa = 3.62KK44 pKa = 11.17GEE46 pKa = 4.05NGTPHH51 pKa = 6.48LQGFVNLKK59 pKa = 10.05KK60 pKa = 10.61KK61 pKa = 9.7MRR63 pKa = 11.84FNPFKK68 pKa = 10.66AALGGRR74 pKa = 11.84AHH76 pKa = 7.3IEE78 pKa = 3.82QARR81 pKa = 11.84GTDD84 pKa = 3.67LDD86 pKa = 3.68NKK88 pKa = 9.98RR89 pKa = 11.84YY90 pKa = 9.18CSKK93 pKa = 11.18GGDD96 pKa = 3.5LLLEE100 pKa = 4.22VGEE103 pKa = 4.64PGKK106 pKa = 10.33QGKK109 pKa = 9.96RR110 pKa = 11.84SDD112 pKa = 3.51LKK114 pKa = 11.13EE115 pKa = 3.9AVTLLNSGANMTAVARR131 pKa = 11.84AYY133 pKa = 9.02PEE135 pKa = 3.65VFIRR139 pKa = 11.84YY140 pKa = 8.65GRR142 pKa = 11.84GLRR145 pKa = 11.84DD146 pKa = 3.45YY147 pKa = 10.83VITAGLSQQRR157 pKa = 11.84AWKK160 pKa = 8.44TEE162 pKa = 3.34VHH164 pKa = 6.42VIVGVPGVGKK174 pKa = 10.03SRR176 pKa = 11.84HH177 pKa = 4.83VSEE180 pKa = 4.27QHH182 pKa = 6.42SDD184 pKa = 3.42IYY186 pKa = 10.2WKK188 pKa = 10.17PRR190 pKa = 11.84GKK192 pKa = 8.98WWDD195 pKa = 4.15GYY197 pKa = 10.73CNQEE201 pKa = 4.39VVCLDD206 pKa = 4.83DD207 pKa = 3.91YY208 pKa = 11.47YY209 pKa = 11.91GWIPYY214 pKa = 10.31DD215 pKa = 4.05DD216 pKa = 5.12LLRR219 pKa = 11.84LCDD222 pKa = 4.13RR223 pKa = 11.84YY224 pKa = 10.18PLRR227 pKa = 11.84VEE229 pKa = 4.36TKK231 pKa = 8.38GGTVSFVAKK240 pKa = 10.32KK241 pKa = 10.3IYY243 pKa = 8.87ITSNKK248 pKa = 8.9QIKK251 pKa = 9.38DD252 pKa = 2.98WYY254 pKa = 10.2NFEE257 pKa = 4.17EE258 pKa = 4.59LKK260 pKa = 10.53VDD262 pKa = 3.39PRR264 pKa = 11.84ALYY267 pKa = 10.43RR268 pKa = 11.84RR269 pKa = 11.84VTSYY273 pKa = 11.31KK274 pKa = 9.81VMRR277 pKa = 11.84EE278 pKa = 3.69GGEE281 pKa = 4.09LYY283 pKa = 10.7DD284 pKa = 4.79VVMTGDD290 pKa = 3.31NKK292 pKa = 10.54INYY295 pKa = 8.4
MM1 pKa = 7.46EE2 pKa = 4.57MAGHH6 pKa = 6.25PCEE9 pKa = 4.97RR10 pKa = 11.84YY11 pKa = 10.13CFTINNYY18 pKa = 9.0SEE20 pKa = 4.06EE21 pKa = 4.62DD22 pKa = 3.4IEE24 pKa = 4.23AVKK27 pKa = 10.96AFLVPDD33 pKa = 3.56NAEE36 pKa = 3.78YY37 pKa = 10.58AIVGKK42 pKa = 10.44EE43 pKa = 3.62KK44 pKa = 11.17GEE46 pKa = 4.05NGTPHH51 pKa = 6.48LQGFVNLKK59 pKa = 10.05KK60 pKa = 10.61KK61 pKa = 9.7MRR63 pKa = 11.84FNPFKK68 pKa = 10.66AALGGRR74 pKa = 11.84AHH76 pKa = 7.3IEE78 pKa = 3.82QARR81 pKa = 11.84GTDD84 pKa = 3.67LDD86 pKa = 3.68NKK88 pKa = 9.98RR89 pKa = 11.84YY90 pKa = 9.18CSKK93 pKa = 11.18GGDD96 pKa = 3.5LLLEE100 pKa = 4.22VGEE103 pKa = 4.64PGKK106 pKa = 10.33QGKK109 pKa = 9.96RR110 pKa = 11.84SDD112 pKa = 3.51LKK114 pKa = 11.13EE115 pKa = 3.9AVTLLNSGANMTAVARR131 pKa = 11.84AYY133 pKa = 9.02PEE135 pKa = 3.65VFIRR139 pKa = 11.84YY140 pKa = 8.65GRR142 pKa = 11.84GLRR145 pKa = 11.84DD146 pKa = 3.45YY147 pKa = 10.83VITAGLSQQRR157 pKa = 11.84AWKK160 pKa = 8.44TEE162 pKa = 3.34VHH164 pKa = 6.42VIVGVPGVGKK174 pKa = 10.03SRR176 pKa = 11.84HH177 pKa = 4.83VSEE180 pKa = 4.27QHH182 pKa = 6.42SDD184 pKa = 3.42IYY186 pKa = 10.2WKK188 pKa = 10.17PRR190 pKa = 11.84GKK192 pKa = 8.98WWDD195 pKa = 4.15GYY197 pKa = 10.73CNQEE201 pKa = 4.39VVCLDD206 pKa = 4.83DD207 pKa = 3.91YY208 pKa = 11.47YY209 pKa = 11.91GWIPYY214 pKa = 10.31DD215 pKa = 4.05DD216 pKa = 5.12LLRR219 pKa = 11.84LCDD222 pKa = 4.13RR223 pKa = 11.84YY224 pKa = 10.18PLRR227 pKa = 11.84VEE229 pKa = 4.36TKK231 pKa = 8.38GGTVSFVAKK240 pKa = 10.32KK241 pKa = 10.3IYY243 pKa = 8.87ITSNKK248 pKa = 8.9QIKK251 pKa = 9.38DD252 pKa = 2.98WYY254 pKa = 10.2NFEE257 pKa = 4.17EE258 pKa = 4.59LKK260 pKa = 10.53VDD262 pKa = 3.39PRR264 pKa = 11.84ALYY267 pKa = 10.43RR268 pKa = 11.84RR269 pKa = 11.84VTSYY273 pKa = 11.31KK274 pKa = 9.81VMRR277 pKa = 11.84EE278 pKa = 3.69GGEE281 pKa = 4.09LYY283 pKa = 10.7DD284 pKa = 4.79VVMTGDD290 pKa = 3.31NKK292 pKa = 10.54INYY295 pKa = 8.4
Molecular weight: 33.69 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|R4L9E4|R4L9E4_9CIRC Capsid protein OS=Bat associated circovirus 1 OX=1868218 GN=cap PE=3 SV=1
MM1 pKa = 7.94PIRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84SRR8 pKa = 11.84YY9 pKa = 7.19SRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84WRR16 pKa = 11.84RR17 pKa = 11.84NTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84VARR26 pKa = 11.84GAYY29 pKa = 8.5RR30 pKa = 11.84WRR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 10.04NGIINVRR41 pKa = 11.84LSATKK46 pKa = 10.02DD47 pKa = 2.63WTMASTTAEE56 pKa = 4.56GYY58 pKa = 9.63NVARR62 pKa = 11.84LEE64 pKa = 4.28VNLRR68 pKa = 11.84QFMPAGPGSAINTKK82 pKa = 10.08SIPWAYY88 pKa = 10.59YY89 pKa = 9.97RR90 pKa = 11.84IRR92 pKa = 11.84KK93 pKa = 7.77MKK95 pKa = 10.71FEE97 pKa = 4.98ILPKK101 pKa = 9.69MIPAQTPYY109 pKa = 10.51RR110 pKa = 11.84YY111 pKa = 10.16GSTAIYY117 pKa = 10.62LGMQAPAPTQGKK129 pKa = 7.78TYY131 pKa = 10.3DD132 pKa = 3.39PHH134 pKa = 7.41LKK136 pKa = 9.95HH137 pKa = 6.48VKK139 pKa = 10.06QNMSGLITDD148 pKa = 3.24QLKK151 pKa = 10.5RR152 pKa = 11.84YY153 pKa = 7.39FTPKK157 pKa = 9.73PDD159 pKa = 3.91LDD161 pKa = 5.18SITSTAWFQPNNKK174 pKa = 9.35ANQVWINMTNDD185 pKa = 3.67NITHH189 pKa = 5.88GQVGWSMEE197 pKa = 4.49RR198 pKa = 11.84ISNMAQNFKK207 pKa = 10.53IRR209 pKa = 11.84VTLYY213 pKa = 8.8VQFRR217 pKa = 11.84EE218 pKa = 4.41FNLIDD223 pKa = 3.88YY224 pKa = 7.74PAQAPLLVDD233 pKa = 4.1EE234 pKa = 5.54EE235 pKa = 4.38PSEE238 pKa = 4.17
MM1 pKa = 7.94PIRR4 pKa = 11.84RR5 pKa = 11.84RR6 pKa = 11.84SRR8 pKa = 11.84YY9 pKa = 7.19SRR11 pKa = 11.84RR12 pKa = 11.84RR13 pKa = 11.84RR14 pKa = 11.84WRR16 pKa = 11.84RR17 pKa = 11.84NTRR20 pKa = 11.84RR21 pKa = 11.84RR22 pKa = 11.84RR23 pKa = 11.84VARR26 pKa = 11.84GAYY29 pKa = 8.5RR30 pKa = 11.84WRR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 10.04NGIINVRR41 pKa = 11.84LSATKK46 pKa = 10.02DD47 pKa = 2.63WTMASTTAEE56 pKa = 4.56GYY58 pKa = 9.63NVARR62 pKa = 11.84LEE64 pKa = 4.28VNLRR68 pKa = 11.84QFMPAGPGSAINTKK82 pKa = 10.08SIPWAYY88 pKa = 10.59YY89 pKa = 9.97RR90 pKa = 11.84IRR92 pKa = 11.84KK93 pKa = 7.77MKK95 pKa = 10.71FEE97 pKa = 4.98ILPKK101 pKa = 9.69MIPAQTPYY109 pKa = 10.51RR110 pKa = 11.84YY111 pKa = 10.16GSTAIYY117 pKa = 10.62LGMQAPAPTQGKK129 pKa = 7.78TYY131 pKa = 10.3DD132 pKa = 3.39PHH134 pKa = 7.41LKK136 pKa = 9.95HH137 pKa = 6.48VKK139 pKa = 10.06QNMSGLITDD148 pKa = 3.24QLKK151 pKa = 10.5RR152 pKa = 11.84YY153 pKa = 7.39FTPKK157 pKa = 9.73PDD159 pKa = 3.91LDD161 pKa = 5.18SITSTAWFQPNNKK174 pKa = 9.35ANQVWINMTNDD185 pKa = 3.67NITHH189 pKa = 5.88GQVGWSMEE197 pKa = 4.49RR198 pKa = 11.84ISNMAQNFKK207 pKa = 10.53IRR209 pKa = 11.84VTLYY213 pKa = 8.8VQFRR217 pKa = 11.84EE218 pKa = 4.41FNLIDD223 pKa = 3.88YY224 pKa = 7.74PAQAPLLVDD233 pKa = 4.1EE234 pKa = 5.54EE235 pKa = 4.38PSEE238 pKa = 4.17
Molecular weight: 27.97 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
533 |
238 |
295 |
266.5 |
30.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.754 ± 0.473 | 1.126 ± 0.659 |
4.878 ± 0.888 | 5.441 ± 1.217 |
2.814 ± 0.074 | 7.317 ± 1.578 |
1.689 ± 0.251 | 5.441 ± 0.75 |
6.942 ± 0.866 | 6.379 ± 0.537 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.002 ± 0.702 | 5.629 ± 0.641 |
5.066 ± 0.97 | 3.752 ± 0.755 |
9.006 ± 1.615 | 4.503 ± 0.562 |
5.441 ± 0.996 | 6.567 ± 1.384 |
2.439 ± 0.294 | 5.816 ± 0.453 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
