
Solanum nodiflorum mottle virus
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Pisoniviricetes; Sobelivirales; Solemoviridae; Sobemovirus
Average proteome isoelectric point is 6.54
Get precalculated fractions of proteins
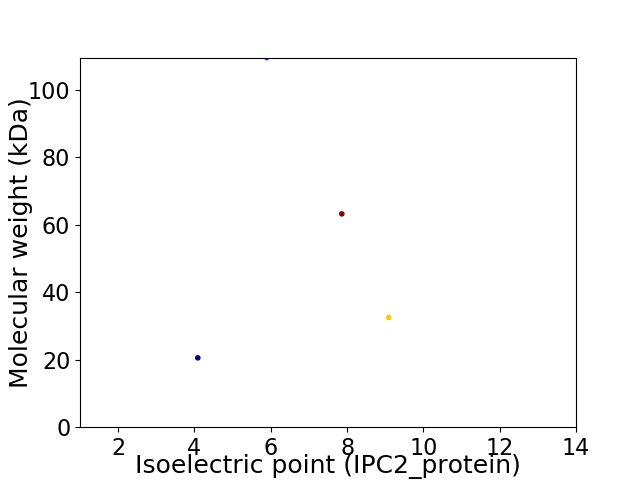
Virtual 2D-PAGE plot for 4 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A1P7XK87|A0A1P7XK87_9VIRU Putative RNA silencing suppressor OS=Solanum nodiflorum mottle virus OX=12471 GN=P1 PE=4 SV=1
MM1 pKa = 7.76PSIILEE7 pKa = 4.51IISSYY12 pKa = 8.42TVNGAPCDD20 pKa = 3.5ISEE23 pKa = 4.61PLPFSRR29 pKa = 11.84EE30 pKa = 4.01LIDD33 pKa = 3.6TKK35 pKa = 11.17KK36 pKa = 10.94LFIDD40 pKa = 4.46LPGDD44 pKa = 3.71NLPDD48 pKa = 3.44SYY50 pKa = 10.92RR51 pKa = 11.84VYY53 pKa = 11.34GRR55 pKa = 11.84FNPACISAVEE65 pKa = 3.82LHH67 pKa = 6.57IVCANCGASYY77 pKa = 10.31WKK79 pKa = 10.2FAEE82 pKa = 4.27LKK84 pKa = 10.25NVEE87 pKa = 4.5LVEE90 pKa = 4.02FDD92 pKa = 3.65YY93 pKa = 11.64VEE95 pKa = 4.92SITRR99 pKa = 11.84PGSLPSVYY107 pKa = 10.07TYY109 pKa = 11.25TNLVDD114 pKa = 4.57AEE116 pKa = 4.46LTVEE120 pKa = 4.13VCDD123 pKa = 4.08EE124 pKa = 4.41SEE126 pKa = 4.28TCSEE130 pKa = 3.99HH131 pKa = 6.86HH132 pKa = 7.12LNTILGLADD141 pKa = 3.34AAGEE145 pKa = 4.02FSLDD149 pKa = 3.48SDD151 pKa = 4.84SDD153 pKa = 4.0EE154 pKa = 4.36EE155 pKa = 4.42DD156 pKa = 3.4QLSLLYY162 pKa = 10.65RR163 pKa = 11.84GDD165 pKa = 3.83LSLEE169 pKa = 3.73RR170 pKa = 11.84DD171 pKa = 3.39AATDD175 pKa = 3.0WRR177 pKa = 11.84HH178 pKa = 6.48RR179 pKa = 11.84EE180 pKa = 3.64TDD182 pKa = 3.37EE183 pKa = 3.99NN184 pKa = 4.01
MM1 pKa = 7.76PSIILEE7 pKa = 4.51IISSYY12 pKa = 8.42TVNGAPCDD20 pKa = 3.5ISEE23 pKa = 4.61PLPFSRR29 pKa = 11.84EE30 pKa = 4.01LIDD33 pKa = 3.6TKK35 pKa = 11.17KK36 pKa = 10.94LFIDD40 pKa = 4.46LPGDD44 pKa = 3.71NLPDD48 pKa = 3.44SYY50 pKa = 10.92RR51 pKa = 11.84VYY53 pKa = 11.34GRR55 pKa = 11.84FNPACISAVEE65 pKa = 3.82LHH67 pKa = 6.57IVCANCGASYY77 pKa = 10.31WKK79 pKa = 10.2FAEE82 pKa = 4.27LKK84 pKa = 10.25NVEE87 pKa = 4.5LVEE90 pKa = 4.02FDD92 pKa = 3.65YY93 pKa = 11.64VEE95 pKa = 4.92SITRR99 pKa = 11.84PGSLPSVYY107 pKa = 10.07TYY109 pKa = 11.25TNLVDD114 pKa = 4.57AEE116 pKa = 4.46LTVEE120 pKa = 4.13VCDD123 pKa = 4.08EE124 pKa = 4.41SEE126 pKa = 4.28TCSEE130 pKa = 3.99HH131 pKa = 6.86HH132 pKa = 7.12LNTILGLADD141 pKa = 3.34AAGEE145 pKa = 4.02FSLDD149 pKa = 3.48SDD151 pKa = 4.84SDD153 pKa = 4.0EE154 pKa = 4.36EE155 pKa = 4.42DD156 pKa = 3.4QLSLLYY162 pKa = 10.65RR163 pKa = 11.84GDD165 pKa = 3.83LSLEE169 pKa = 3.73RR170 pKa = 11.84DD171 pKa = 3.39AATDD175 pKa = 3.0WRR177 pKa = 11.84HH178 pKa = 6.48RR179 pKa = 11.84EE180 pKa = 3.64TDD182 pKa = 3.37EE183 pKa = 3.99NN184 pKa = 4.01
Molecular weight: 20.56 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A1P7XK78|A0A1P7XK78_9VIRU N-terminal protein OS=Solanum nodiflorum mottle virus OX=12471 PE=4 SV=1
MM1 pKa = 7.1VKK3 pKa = 10.0RR4 pKa = 11.84RR5 pKa = 11.84PRR7 pKa = 11.84LRR9 pKa = 11.84QLTVMEE15 pKa = 4.7RR16 pKa = 11.84VTEE19 pKa = 4.14RR20 pKa = 11.84NPPARR25 pKa = 11.84KK26 pKa = 8.95SRR28 pKa = 11.84PRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 9.8GGQTTMVTAPIAGTMVYY51 pKa = 9.94RR52 pKa = 11.84RR53 pKa = 11.84NPILMNGRR61 pKa = 11.84RR62 pKa = 11.84GITVCHH68 pKa = 6.23SEE70 pKa = 4.2VVLAVQSTATAFSATSVTLAPNTFSWLSVMGALFSKK106 pKa = 9.43WRR108 pKa = 11.84WISLRR113 pKa = 11.84ATYY116 pKa = 10.53VPEE119 pKa = 4.29TATTTPGIVAMAFQYY134 pKa = 11.43DD135 pKa = 3.8NTDD138 pKa = 3.45TLPVGNAGMSSLYY151 pKa = 10.31GYY153 pKa = 10.95VSGAPWAGFEE163 pKa = 4.18GSKK166 pKa = 10.88LLTEE170 pKa = 4.86KK171 pKa = 9.26PTTPIPSGAIATQLDD186 pKa = 4.13CTNFGLKK193 pKa = 8.93WYY195 pKa = 9.51QYY197 pKa = 9.43KK198 pKa = 10.7AVMPAGDD205 pKa = 4.11SGNLYY210 pKa = 10.51VPAQLIVGNLGANNPLRR227 pKa = 11.84YY228 pKa = 10.01GEE230 pKa = 4.01VHH232 pKa = 5.56IQYY235 pKa = 9.13EE236 pKa = 4.48VEE238 pKa = 4.7FIEE241 pKa = 5.57PIPPTSNALRR251 pKa = 11.84SVTDD255 pKa = 4.06EE256 pKa = 3.44ILKK259 pKa = 9.98RR260 pKa = 11.84EE261 pKa = 3.83RR262 pKa = 11.84RR263 pKa = 11.84EE264 pKa = 3.9LCVNEE269 pKa = 4.14PPAVVVNTEE278 pKa = 3.63VLLPSKK284 pKa = 9.29EE285 pKa = 4.35TEE287 pKa = 3.43IDD289 pKa = 3.36KK290 pKa = 11.58AEE292 pKa = 4.96LIPP295 pKa = 5.04
MM1 pKa = 7.1VKK3 pKa = 10.0RR4 pKa = 11.84RR5 pKa = 11.84PRR7 pKa = 11.84LRR9 pKa = 11.84QLTVMEE15 pKa = 4.7RR16 pKa = 11.84VTEE19 pKa = 4.14RR20 pKa = 11.84NPPARR25 pKa = 11.84KK26 pKa = 8.95SRR28 pKa = 11.84PRR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 9.8GGQTTMVTAPIAGTMVYY51 pKa = 9.94RR52 pKa = 11.84RR53 pKa = 11.84NPILMNGRR61 pKa = 11.84RR62 pKa = 11.84GITVCHH68 pKa = 6.23SEE70 pKa = 4.2VVLAVQSTATAFSATSVTLAPNTFSWLSVMGALFSKK106 pKa = 9.43WRR108 pKa = 11.84WISLRR113 pKa = 11.84ATYY116 pKa = 10.53VPEE119 pKa = 4.29TATTTPGIVAMAFQYY134 pKa = 11.43DD135 pKa = 3.8NTDD138 pKa = 3.45TLPVGNAGMSSLYY151 pKa = 10.31GYY153 pKa = 10.95VSGAPWAGFEE163 pKa = 4.18GSKK166 pKa = 10.88LLTEE170 pKa = 4.86KK171 pKa = 9.26PTTPIPSGAIATQLDD186 pKa = 4.13CTNFGLKK193 pKa = 8.93WYY195 pKa = 9.51QYY197 pKa = 9.43KK198 pKa = 10.7AVMPAGDD205 pKa = 4.11SGNLYY210 pKa = 10.51VPAQLIVGNLGANNPLRR227 pKa = 11.84YY228 pKa = 10.01GEE230 pKa = 4.01VHH232 pKa = 5.56IQYY235 pKa = 9.13EE236 pKa = 4.48VEE238 pKa = 4.7FIEE241 pKa = 5.57PIPPTSNALRR251 pKa = 11.84SVTDD255 pKa = 4.06EE256 pKa = 3.44ILKK259 pKa = 9.98RR260 pKa = 11.84EE261 pKa = 3.83RR262 pKa = 11.84RR263 pKa = 11.84EE264 pKa = 3.9LCVNEE269 pKa = 4.14PPAVVVNTEE278 pKa = 3.63VLLPSKK284 pKa = 9.29EE285 pKa = 4.35TEE287 pKa = 3.43IDD289 pKa = 3.36KK290 pKa = 11.58AEE292 pKa = 4.96LIPP295 pKa = 5.04
Molecular weight: 32.54 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2039 |
184 |
981 |
509.8 |
56.49 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.376 ± 0.461 | 1.717 ± 0.27 |
4.561 ± 0.664 | 7.504 ± 0.516 |
3.041 ± 0.344 | 7.013 ± 0.521 |
1.864 ± 0.388 | 4.512 ± 0.197 |
5.542 ± 0.602 | 10.152 ± 0.412 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.109 ± 0.248 | 3.335 ± 0.359 |
6.13 ± 0.504 | 2.501 ± 0.235 |
5.64 ± 0.424 | 10.495 ± 1.191 |
5.444 ± 0.91 | 7.013 ± 0.462 |
1.913 ± 0.326 | 3.139 ± 0.166 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
