
Gordonia phage Tiamoceli
Taxonomy: Viruses; Duplodnaviria; Heunggongvirae; Uroviricota; Caudoviricetes; Caudovirales; Siphoviridae; unclassified Siphoviridae
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
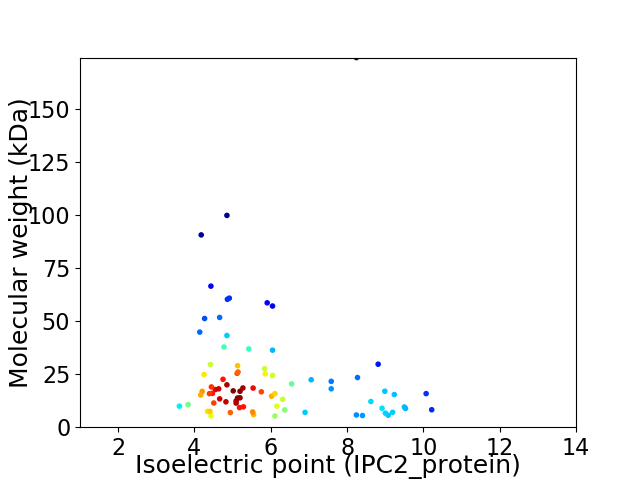
Virtual 2D-PAGE plot for 80 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A411CSG1|A0A411CSG1_9CAUD Uncharacterized protein OS=Gordonia phage Tiamoceli OX=2510508 GN=41 PE=4 SV=1
MM1 pKa = 7.54TSPPGGGTPIVPDD14 pKa = 3.62KK15 pKa = 11.06GYY17 pKa = 8.24TAQTIYY23 pKa = 10.74QLQNVDD29 pKa = 3.54PDD31 pKa = 3.92ALGAEE36 pKa = 4.47ANLDD40 pKa = 3.39LWEE43 pKa = 4.1MLEE46 pKa = 3.95NFRR49 pKa = 11.84NKK51 pKa = 10.5LLTDD55 pKa = 4.12LLGGFASVPIAILDD69 pKa = 4.5GIGDD73 pKa = 4.34VISAITGILNGDD85 pKa = 3.68FDD87 pKa = 6.02DD88 pKa = 4.15LHH90 pKa = 6.7EE91 pKa = 4.28WAQNVIDD98 pKa = 4.6GFGNLLDD105 pKa = 4.57ALQGNYY111 pKa = 9.45TGTDD115 pKa = 3.03AFLLQIQTWAGDD127 pKa = 3.59LFAWVQDD134 pKa = 3.58VADD137 pKa = 4.9FVQNLLDD144 pKa = 4.95AILRR148 pKa = 11.84GIRR151 pKa = 11.84GIPVVGGAIADD162 pKa = 4.14VISDD166 pKa = 3.73LTGLNTKK173 pKa = 8.22ATNALSTANSAAADD187 pKa = 3.42AAAAQVAINDD197 pKa = 4.34LSADD201 pKa = 3.54MAALQVTQVYY211 pKa = 10.05EE212 pKa = 4.17SLSQHH217 pKa = 6.94DD218 pKa = 3.7IVSFPRR224 pKa = 11.84YY225 pKa = 7.89TLGLGADD232 pKa = 4.09GTTGSAGSMSCADD245 pKa = 3.75ASHH248 pKa = 5.99SHH250 pKa = 6.33SSHH253 pKa = 5.18SHH255 pKa = 4.71SVGHH259 pKa = 6.16EE260 pKa = 3.92MPTANPGKK268 pKa = 10.17GVVGYY273 pKa = 10.78VPLFVNRR280 pKa = 11.84FCRR283 pKa = 11.84PRR285 pKa = 11.84RR286 pKa = 11.84LKK288 pKa = 10.88LITGSTGWSIFSIDD302 pKa = 3.32YY303 pKa = 9.29WYY305 pKa = 11.23VALCVYY311 pKa = 10.29NPNTGNVEE319 pKa = 4.2KK320 pKa = 10.78VWDD323 pKa = 4.22GGDD326 pKa = 3.2QKK328 pKa = 11.65GAITTAAKK336 pKa = 10.29LHH338 pKa = 6.16AFDD341 pKa = 4.97MGTLNDD347 pKa = 3.89VSPGTILFGAQLQNAGLIVNTRR369 pKa = 11.84PIAALWQPGLQDD381 pKa = 3.77PSAEE385 pKa = 4.31LLTAPLYY392 pKa = 10.48NLSGQSSIPSSVPLSSLTPNNDD414 pKa = 3.09TLPWMGVGVDD424 pKa = 3.85PVV426 pKa = 3.09
MM1 pKa = 7.54TSPPGGGTPIVPDD14 pKa = 3.62KK15 pKa = 11.06GYY17 pKa = 8.24TAQTIYY23 pKa = 10.74QLQNVDD29 pKa = 3.54PDD31 pKa = 3.92ALGAEE36 pKa = 4.47ANLDD40 pKa = 3.39LWEE43 pKa = 4.1MLEE46 pKa = 3.95NFRR49 pKa = 11.84NKK51 pKa = 10.5LLTDD55 pKa = 4.12LLGGFASVPIAILDD69 pKa = 4.5GIGDD73 pKa = 4.34VISAITGILNGDD85 pKa = 3.68FDD87 pKa = 6.02DD88 pKa = 4.15LHH90 pKa = 6.7EE91 pKa = 4.28WAQNVIDD98 pKa = 4.6GFGNLLDD105 pKa = 4.57ALQGNYY111 pKa = 9.45TGTDD115 pKa = 3.03AFLLQIQTWAGDD127 pKa = 3.59LFAWVQDD134 pKa = 3.58VADD137 pKa = 4.9FVQNLLDD144 pKa = 4.95AILRR148 pKa = 11.84GIRR151 pKa = 11.84GIPVVGGAIADD162 pKa = 4.14VISDD166 pKa = 3.73LTGLNTKK173 pKa = 8.22ATNALSTANSAAADD187 pKa = 3.42AAAAQVAINDD197 pKa = 4.34LSADD201 pKa = 3.54MAALQVTQVYY211 pKa = 10.05EE212 pKa = 4.17SLSQHH217 pKa = 6.94DD218 pKa = 3.7IVSFPRR224 pKa = 11.84YY225 pKa = 7.89TLGLGADD232 pKa = 4.09GTTGSAGSMSCADD245 pKa = 3.75ASHH248 pKa = 5.99SHH250 pKa = 6.33SSHH253 pKa = 5.18SHH255 pKa = 4.71SVGHH259 pKa = 6.16EE260 pKa = 3.92MPTANPGKK268 pKa = 10.17GVVGYY273 pKa = 10.78VPLFVNRR280 pKa = 11.84FCRR283 pKa = 11.84PRR285 pKa = 11.84RR286 pKa = 11.84LKK288 pKa = 10.88LITGSTGWSIFSIDD302 pKa = 3.32YY303 pKa = 9.29WYY305 pKa = 11.23VALCVYY311 pKa = 10.29NPNTGNVEE319 pKa = 4.2KK320 pKa = 10.78VWDD323 pKa = 4.22GGDD326 pKa = 3.2QKK328 pKa = 11.65GAITTAAKK336 pKa = 10.29LHH338 pKa = 6.16AFDD341 pKa = 4.97MGTLNDD347 pKa = 3.89VSPGTILFGAQLQNAGLIVNTRR369 pKa = 11.84PIAALWQPGLQDD381 pKa = 3.77PSAEE385 pKa = 4.31LLTAPLYY392 pKa = 10.48NLSGQSSIPSSVPLSSLTPNNDD414 pKa = 3.09TLPWMGVGVDD424 pKa = 3.85PVV426 pKa = 3.09
Molecular weight: 44.77 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A411CSI3|A0A411CSI3_9CAUD Uncharacterized protein OS=Gordonia phage Tiamoceli OX=2510508 GN=74 PE=4 SV=1
MM1 pKa = 7.78AEE3 pKa = 4.33KK4 pKa = 10.64LHH6 pKa = 6.49DD7 pKa = 4.07LRR9 pKa = 11.84SFAEE13 pKa = 4.27VAGVKK18 pKa = 8.49YY19 pKa = 7.12TTMRR23 pKa = 11.84RR24 pKa = 11.84YY25 pKa = 9.56HH26 pKa = 5.45ATATKK31 pKa = 10.21RR32 pKa = 11.84RR33 pKa = 11.84AEE35 pKa = 3.96AAADD39 pKa = 3.85PEE41 pKa = 4.68VKK43 pKa = 10.5LPAWLIPPPDD53 pKa = 3.87DD54 pKa = 4.52RR55 pKa = 11.84VGQSPVWRR63 pKa = 11.84DD64 pKa = 3.03RR65 pKa = 11.84TVRR68 pKa = 11.84KK69 pKa = 9.15WIEE72 pKa = 3.77SRR74 pKa = 11.84PRR76 pKa = 11.84AATHH80 pKa = 5.72ATTT83 pKa = 4.42
MM1 pKa = 7.78AEE3 pKa = 4.33KK4 pKa = 10.64LHH6 pKa = 6.49DD7 pKa = 4.07LRR9 pKa = 11.84SFAEE13 pKa = 4.27VAGVKK18 pKa = 8.49YY19 pKa = 7.12TTMRR23 pKa = 11.84RR24 pKa = 11.84YY25 pKa = 9.56HH26 pKa = 5.45ATATKK31 pKa = 10.21RR32 pKa = 11.84RR33 pKa = 11.84AEE35 pKa = 3.96AAADD39 pKa = 3.85PEE41 pKa = 4.68VKK43 pKa = 10.5LPAWLIPPPDD53 pKa = 3.87DD54 pKa = 4.52RR55 pKa = 11.84VGQSPVWRR63 pKa = 11.84DD64 pKa = 3.03RR65 pKa = 11.84TVRR68 pKa = 11.84KK69 pKa = 9.15WIEE72 pKa = 3.77SRR74 pKa = 11.84PRR76 pKa = 11.84AATHH80 pKa = 5.72ATTT83 pKa = 4.42
Molecular weight: 9.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
17862 |
47 |
1705 |
223.3 |
24.15 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.658 ± 0.408 | 0.795 ± 0.105 |
7.272 ± 0.419 | 5.682 ± 0.353 |
2.581 ± 0.201 | 8.958 ± 0.375 |
1.814 ± 0.165 | 4.496 ± 0.19 |
2.9 ± 0.214 | 7.687 ± 0.252 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.909 ± 0.106 | 2.794 ± 0.176 |
5.985 ± 0.259 | 3.79 ± 0.146 |
6.931 ± 0.394 | 4.938 ± 0.278 |
7.16 ± 0.284 | 7.172 ± 0.219 |
2.155 ± 0.152 | 2.323 ± 0.203 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
