
Saccharomyces cerevisiae killer virus M1 (ScV-M1) (Saccharomyces cerevisiae virus M1)
Taxonomy: Viruses; Riboviria; dsRNA viruses; unclassified dsRNA viruses
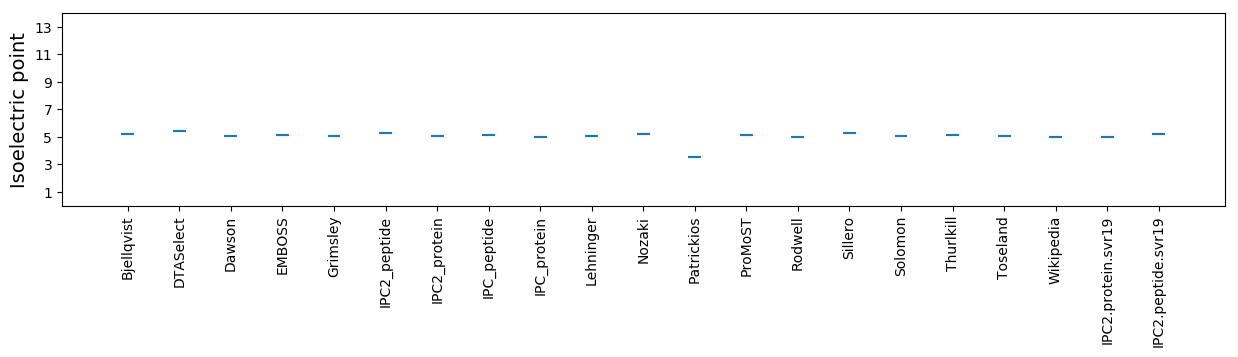
Average proteome isoelectric point is 4.97
Get precalculated fractions of proteins
Virtual 2D-PAGE plot for 1 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P01546|M11P_SCVM1 M1-1 protoxin OS=Saccharomyces cerevisiae killer virus M1 OX=12450 PE=2 SV=1
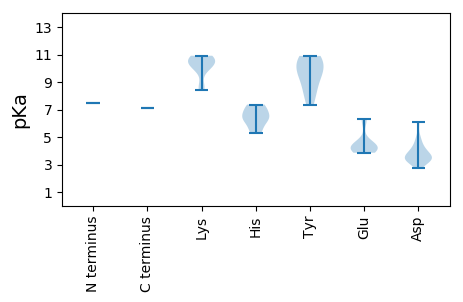
MM1 pKa = 7.48TKK3 pKa = 8.67PTQVLVRR10 pKa = 11.84SVSILFFITLLHH22 pKa = 6.4LVVALNDD29 pKa = 3.45VAGPAEE35 pKa = 4.1TAPVSLLPRR44 pKa = 11.84EE45 pKa = 4.38APWYY49 pKa = 9.68DD50 pKa = 3.92KK51 pKa = 10.62IWEE54 pKa = 4.26VKK56 pKa = 10.09DD57 pKa = 3.27WLLQRR62 pKa = 11.84ATDD65 pKa = 4.23GNWGKK70 pKa = 10.91SITWGSFVASDD81 pKa = 3.42AGVVIFGINVCKK93 pKa = 10.82NCVGEE98 pKa = 4.21RR99 pKa = 11.84KK100 pKa = 10.0DD101 pKa = 6.11DD102 pKa = 3.4ISTDD106 pKa = 3.5CGKK109 pKa = 8.45QTLALLVSIFVAVTSGHH126 pKa = 5.53HH127 pKa = 6.46LIWGGNRR134 pKa = 11.84PVSQSDD140 pKa = 3.6PNGATVARR148 pKa = 11.84RR149 pKa = 11.84DD150 pKa = 3.29ISTVADD156 pKa = 3.53GDD158 pKa = 3.87IPLDD162 pKa = 4.16FSALNDD168 pKa = 3.43ILNEE172 pKa = 4.0HH173 pKa = 7.36GISILPANASQYY185 pKa = 10.4VKK187 pKa = 10.68RR188 pKa = 11.84SDD190 pKa = 3.62TAEE193 pKa = 3.96HH194 pKa = 5.31TTSFVVTNNYY204 pKa = 8.38TSLHH208 pKa = 6.27TDD210 pKa = 4.83LIHH213 pKa = 7.0HH214 pKa = 6.78GNGTYY219 pKa = 7.36TTFTTPHH226 pKa = 6.16IPAVAKK232 pKa = 10.11RR233 pKa = 11.84YY234 pKa = 8.43VYY236 pKa = 9.89PMCEE240 pKa = 3.86HH241 pKa = 7.32GIKK244 pKa = 10.36ASYY247 pKa = 10.83CMALNDD253 pKa = 4.87AMVSANGNLYY263 pKa = 10.73GLAEE267 pKa = 4.24KK268 pKa = 10.65LFSEE272 pKa = 6.28DD273 pKa = 2.77EE274 pKa = 4.6GQWEE278 pKa = 4.1TNYY281 pKa = 10.9YY282 pKa = 9.33KK283 pKa = 10.82LYY285 pKa = 9.74WSTGQWIMSMKK296 pKa = 10.33FIEE299 pKa = 4.33EE300 pKa = 4.46SIDD303 pKa = 3.31NANNDD308 pKa = 4.21FEE310 pKa = 6.26GCDD313 pKa = 3.28TGHH316 pKa = 7.08
MM1 pKa = 7.48TKK3 pKa = 8.67PTQVLVRR10 pKa = 11.84SVSILFFITLLHH22 pKa = 6.4LVVALNDD29 pKa = 3.45VAGPAEE35 pKa = 4.1TAPVSLLPRR44 pKa = 11.84EE45 pKa = 4.38APWYY49 pKa = 9.68DD50 pKa = 3.92KK51 pKa = 10.62IWEE54 pKa = 4.26VKK56 pKa = 10.09DD57 pKa = 3.27WLLQRR62 pKa = 11.84ATDD65 pKa = 4.23GNWGKK70 pKa = 10.91SITWGSFVASDD81 pKa = 3.42AGVVIFGINVCKK93 pKa = 10.82NCVGEE98 pKa = 4.21RR99 pKa = 11.84KK100 pKa = 10.0DD101 pKa = 6.11DD102 pKa = 3.4ISTDD106 pKa = 3.5CGKK109 pKa = 8.45QTLALLVSIFVAVTSGHH126 pKa = 5.53HH127 pKa = 6.46LIWGGNRR134 pKa = 11.84PVSQSDD140 pKa = 3.6PNGATVARR148 pKa = 11.84RR149 pKa = 11.84DD150 pKa = 3.29ISTVADD156 pKa = 3.53GDD158 pKa = 3.87IPLDD162 pKa = 4.16FSALNDD168 pKa = 3.43ILNEE172 pKa = 4.0HH173 pKa = 7.36GISILPANASQYY185 pKa = 10.4VKK187 pKa = 10.68RR188 pKa = 11.84SDD190 pKa = 3.62TAEE193 pKa = 3.96HH194 pKa = 5.31TTSFVVTNNYY204 pKa = 8.38TSLHH208 pKa = 6.27TDD210 pKa = 4.83LIHH213 pKa = 7.0HH214 pKa = 6.78GNGTYY219 pKa = 7.36TTFTTPHH226 pKa = 6.16IPAVAKK232 pKa = 10.11RR233 pKa = 11.84YY234 pKa = 8.43VYY236 pKa = 9.89PMCEE240 pKa = 3.86HH241 pKa = 7.32GIKK244 pKa = 10.36ASYY247 pKa = 10.83CMALNDD253 pKa = 4.87AMVSANGNLYY263 pKa = 10.73GLAEE267 pKa = 4.24KK268 pKa = 10.65LFSEE272 pKa = 6.28DD273 pKa = 2.77EE274 pKa = 4.6GQWEE278 pKa = 4.1TNYY281 pKa = 10.9YY282 pKa = 9.33KK283 pKa = 10.82LYY285 pKa = 9.74WSTGQWIMSMKK296 pKa = 10.33FIEE299 pKa = 4.33EE300 pKa = 4.46SIDD303 pKa = 3.31NANNDD308 pKa = 4.21FEE310 pKa = 6.26GCDD313 pKa = 3.28TGHH316 pKa = 7.08
Molecular weight: 34.83 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P01546|M11P_SCVM1 M1-1 protoxin OS=Saccharomyces cerevisiae killer virus M1 OX=12450 PE=2 SV=1
MM1 pKa = 7.48TKK3 pKa = 8.67PTQVLVRR10 pKa = 11.84SVSILFFITLLHH22 pKa = 6.4LVVALNDD29 pKa = 3.45VAGPAEE35 pKa = 4.1TAPVSLLPRR44 pKa = 11.84EE45 pKa = 4.38APWYY49 pKa = 9.68DD50 pKa = 3.92KK51 pKa = 10.62IWEE54 pKa = 4.26VKK56 pKa = 10.09DD57 pKa = 3.27WLLQRR62 pKa = 11.84ATDD65 pKa = 4.23GNWGKK70 pKa = 10.91SITWGSFVASDD81 pKa = 3.42AGVVIFGINVCKK93 pKa = 10.82NCVGEE98 pKa = 4.21RR99 pKa = 11.84KK100 pKa = 10.0DD101 pKa = 6.11DD102 pKa = 3.4ISTDD106 pKa = 3.5CGKK109 pKa = 8.45QTLALLVSIFVAVTSGHH126 pKa = 5.53HH127 pKa = 6.46LIWGGNRR134 pKa = 11.84PVSQSDD140 pKa = 3.6PNGATVARR148 pKa = 11.84RR149 pKa = 11.84DD150 pKa = 3.29ISTVADD156 pKa = 3.53GDD158 pKa = 3.87IPLDD162 pKa = 4.16FSALNDD168 pKa = 3.43ILNEE172 pKa = 4.0HH173 pKa = 7.36GISILPANASQYY185 pKa = 10.4VKK187 pKa = 10.68RR188 pKa = 11.84SDD190 pKa = 3.62TAEE193 pKa = 3.96HH194 pKa = 5.31TTSFVVTNNYY204 pKa = 8.38TSLHH208 pKa = 6.27TDD210 pKa = 4.83LIHH213 pKa = 7.0HH214 pKa = 6.78GNGTYY219 pKa = 7.36TTFTTPHH226 pKa = 6.16IPAVAKK232 pKa = 10.11RR233 pKa = 11.84YY234 pKa = 8.43VYY236 pKa = 9.89PMCEE240 pKa = 3.86HH241 pKa = 7.32GIKK244 pKa = 10.36ASYY247 pKa = 10.83CMALNDD253 pKa = 4.87AMVSANGNLYY263 pKa = 10.73GLAEE267 pKa = 4.24KK268 pKa = 10.65LFSEE272 pKa = 6.28DD273 pKa = 2.77EE274 pKa = 4.6GQWEE278 pKa = 4.1TNYY281 pKa = 10.9YY282 pKa = 9.33KK283 pKa = 10.82LYY285 pKa = 9.74WSTGQWIMSMKK296 pKa = 10.33FIEE299 pKa = 4.33EE300 pKa = 4.46SIDD303 pKa = 3.31NANNDD308 pKa = 4.21FEE310 pKa = 6.26GCDD313 pKa = 3.28TGHH316 pKa = 7.08
MM1 pKa = 7.48TKK3 pKa = 8.67PTQVLVRR10 pKa = 11.84SVSILFFITLLHH22 pKa = 6.4LVVALNDD29 pKa = 3.45VAGPAEE35 pKa = 4.1TAPVSLLPRR44 pKa = 11.84EE45 pKa = 4.38APWYY49 pKa = 9.68DD50 pKa = 3.92KK51 pKa = 10.62IWEE54 pKa = 4.26VKK56 pKa = 10.09DD57 pKa = 3.27WLLQRR62 pKa = 11.84ATDD65 pKa = 4.23GNWGKK70 pKa = 10.91SITWGSFVASDD81 pKa = 3.42AGVVIFGINVCKK93 pKa = 10.82NCVGEE98 pKa = 4.21RR99 pKa = 11.84KK100 pKa = 10.0DD101 pKa = 6.11DD102 pKa = 3.4ISTDD106 pKa = 3.5CGKK109 pKa = 8.45QTLALLVSIFVAVTSGHH126 pKa = 5.53HH127 pKa = 6.46LIWGGNRR134 pKa = 11.84PVSQSDD140 pKa = 3.6PNGATVARR148 pKa = 11.84RR149 pKa = 11.84DD150 pKa = 3.29ISTVADD156 pKa = 3.53GDD158 pKa = 3.87IPLDD162 pKa = 4.16FSALNDD168 pKa = 3.43ILNEE172 pKa = 4.0HH173 pKa = 7.36GISILPANASQYY185 pKa = 10.4VKK187 pKa = 10.68RR188 pKa = 11.84SDD190 pKa = 3.62TAEE193 pKa = 3.96HH194 pKa = 5.31TTSFVVTNNYY204 pKa = 8.38TSLHH208 pKa = 6.27TDD210 pKa = 4.83LIHH213 pKa = 7.0HH214 pKa = 6.78GNGTYY219 pKa = 7.36TTFTTPHH226 pKa = 6.16IPAVAKK232 pKa = 10.11RR233 pKa = 11.84YY234 pKa = 8.43VYY236 pKa = 9.89PMCEE240 pKa = 3.86HH241 pKa = 7.32GIKK244 pKa = 10.36ASYY247 pKa = 10.83CMALNDD253 pKa = 4.87AMVSANGNLYY263 pKa = 10.73GLAEE267 pKa = 4.24KK268 pKa = 10.65LFSEE272 pKa = 6.28DD273 pKa = 2.77EE274 pKa = 4.6GQWEE278 pKa = 4.1TNYY281 pKa = 10.9YY282 pKa = 9.33KK283 pKa = 10.82LYY285 pKa = 9.74WSTGQWIMSMKK296 pKa = 10.33FIEE299 pKa = 4.33EE300 pKa = 4.46SIDD303 pKa = 3.31NANNDD308 pKa = 4.21FEE310 pKa = 6.26GCDD313 pKa = 3.28TGHH316 pKa = 7.08
Molecular weight: 34.83 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
316 |
316 |
316 |
316.0 |
34.83 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.911 ± 0.0 | 1.899 ± 0.0 |
6.646 ± 0.0 | 4.43 ± 0.0 |
3.481 ± 0.0 | 7.278 ± 0.0 |
3.481 ± 0.0 | 6.329 ± 0.0 |
4.114 ± 0.0 | 7.911 ± 0.0 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.899 ± 0.0 | 6.013 ± 0.0 |
3.797 ± 0.0 | 2.215 ± 0.0 |
2.848 ± 0.0 | 7.595 ± 0.0 |
7.911 ± 0.0 | 7.911 ± 0.0 |
2.848 ± 0.0 | 3.481 ± 0.0 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
