
Barley stripe mosaic virus (BSMV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Hordeivirus
Average proteome isoelectric point is 6.52
Get precalculated fractions of proteins
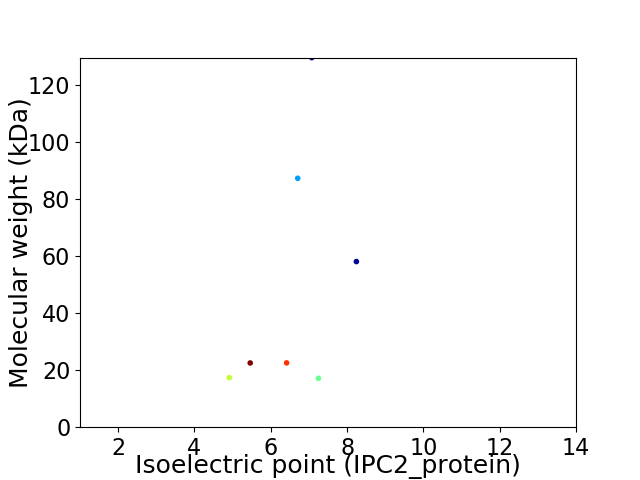
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P04869|TGB2_BSMV Movement protein TGB2 OS=Barley stripe mosaic virus OX=12327 PE=1 SV=2
MM1 pKa = 7.68AMPHH5 pKa = 6.79PLEE8 pKa = 4.7CCCPQCLPSSEE19 pKa = 4.51SFPIYY24 pKa = 10.46GEE26 pKa = 3.92QEE28 pKa = 4.07IPCSEE33 pKa = 4.28TQAEE37 pKa = 4.35TTPVEE42 pKa = 3.99KK43 pKa = 9.75TVRR46 pKa = 11.84ANVLTDD52 pKa = 3.17ILDD55 pKa = 3.53DD56 pKa = 3.99HH57 pKa = 7.29YY58 pKa = 11.73YY59 pKa = 10.97AILASLFIIALWLLYY74 pKa = 9.78IYY76 pKa = 10.4LSSIPTEE83 pKa = 4.04TGPYY87 pKa = 9.81FYY89 pKa = 10.8QDD91 pKa = 3.47LNSVKK96 pKa = 9.98IYY98 pKa = 10.72GIGATNPEE106 pKa = 4.48VIAAIHH112 pKa = 6.19HH113 pKa = 4.96WQKK116 pKa = 11.06YY117 pKa = 8.24PFGEE121 pKa = 4.13SPMWGGLISVLSILLKK137 pKa = 10.58PLTLVFALSFFLLLSSKK154 pKa = 10.48RR155 pKa = 3.67
MM1 pKa = 7.68AMPHH5 pKa = 6.79PLEE8 pKa = 4.7CCCPQCLPSSEE19 pKa = 4.51SFPIYY24 pKa = 10.46GEE26 pKa = 3.92QEE28 pKa = 4.07IPCSEE33 pKa = 4.28TQAEE37 pKa = 4.35TTPVEE42 pKa = 3.99KK43 pKa = 9.75TVRR46 pKa = 11.84ANVLTDD52 pKa = 3.17ILDD55 pKa = 3.53DD56 pKa = 3.99HH57 pKa = 7.29YY58 pKa = 11.73YY59 pKa = 10.97AILASLFIIALWLLYY74 pKa = 9.78IYY76 pKa = 10.4LSSIPTEE83 pKa = 4.04TGPYY87 pKa = 9.81FYY89 pKa = 10.8QDD91 pKa = 3.47LNSVKK96 pKa = 9.98IYY98 pKa = 10.72GIGATNPEE106 pKa = 4.48VIAAIHH112 pKa = 6.19HH113 pKa = 4.96WQKK116 pKa = 11.06YY117 pKa = 8.24PFGEE121 pKa = 4.13SPMWGGLISVLSILLKK137 pKa = 10.58PLTLVFALSFFLLLSSKK154 pKa = 10.48RR155 pKa = 3.67
Molecular weight: 17.38 kDa
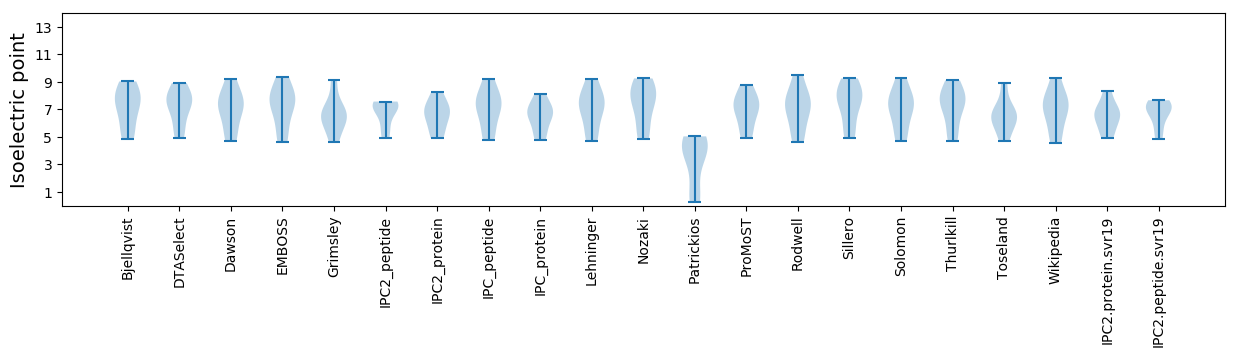
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P04868|TGB3_BSMV Movement protein TGB3 OS=Barley stripe mosaic virus OX=12327 PE=1 SV=1
MM1 pKa = 8.03DD2 pKa = 3.25MTKK5 pKa = 9.39TVEE8 pKa = 4.07EE9 pKa = 4.57KK10 pKa = 9.56KK11 pKa = 9.43TNGTDD16 pKa = 3.15SVKK19 pKa = 10.76GVFEE23 pKa = 4.28NSTIPKK29 pKa = 9.52VPTGQEE35 pKa = 3.63MGGDD39 pKa = 3.74GSSTSKK45 pKa = 10.8LKK47 pKa = 9.43EE48 pKa = 3.98TLKK51 pKa = 11.09VADD54 pKa = 3.99QTPLSVDD61 pKa = 2.98NGAKK65 pKa = 10.12SKK67 pKa = 10.84LDD69 pKa = 3.34SSDD72 pKa = 3.38RR73 pKa = 11.84QVPGVADD80 pKa = 3.68QTPLSVDD87 pKa = 2.98NGAKK91 pKa = 10.12SKK93 pKa = 10.84LDD95 pKa = 3.34SSDD98 pKa = 3.39RR99 pKa = 11.84QVPGPEE105 pKa = 4.03LKK107 pKa = 10.8PNVKK111 pKa = 9.88KK112 pKa = 10.8SKK114 pKa = 10.08KK115 pKa = 10.03KK116 pKa = 10.41RR117 pKa = 11.84IQKK120 pKa = 8.88PAQPSGPNDD129 pKa = 3.45LKK131 pKa = 11.36GGTKK135 pKa = 10.28GSSQVGEE142 pKa = 4.19NVSEE146 pKa = 4.21NYY148 pKa = 9.55TGISKK153 pKa = 10.04EE154 pKa = 3.99AAKK157 pKa = 10.32QKK159 pKa = 10.65QKK161 pKa = 9.58TPKK164 pKa = 8.97SVKK167 pKa = 9.0MQSNLADD174 pKa = 3.36KK175 pKa = 10.72FKK177 pKa = 11.41ANDD180 pKa = 3.36TRR182 pKa = 11.84RR183 pKa = 11.84SEE185 pKa = 4.85LINKK189 pKa = 7.42FQQFVHH195 pKa = 5.52EE196 pKa = 4.59TCLKK200 pKa = 10.69SDD202 pKa = 3.89FEE204 pKa = 4.61YY205 pKa = 10.4TGRR208 pKa = 11.84QYY210 pKa = 10.96FRR212 pKa = 11.84ARR214 pKa = 11.84SNFFEE219 pKa = 4.31MIKK222 pKa = 10.44LASLYY227 pKa = 10.48DD228 pKa = 3.56KK229 pKa = 10.82HH230 pKa = 8.26LKK232 pKa = 9.93EE233 pKa = 4.47CMARR237 pKa = 11.84ACTLEE242 pKa = 3.76RR243 pKa = 11.84EE244 pKa = 4.05RR245 pKa = 11.84LKK247 pKa = 10.99RR248 pKa = 11.84KK249 pKa = 9.57LLLVRR254 pKa = 11.84ALKK257 pKa = 10.01PAVDD261 pKa = 4.64FLTGIISGVPGSGKK275 pKa = 8.33STIVRR280 pKa = 11.84TLLKK284 pKa = 10.86GEE286 pKa = 4.48FPAVCALANPALMNDD301 pKa = 3.38YY302 pKa = 11.01SGIEE306 pKa = 4.06GVYY309 pKa = 10.94GLDD312 pKa = 4.2DD313 pKa = 4.69LLLSAVPITSDD324 pKa = 3.63LLIIDD329 pKa = 5.26EE330 pKa = 4.37YY331 pKa = 10.97TLAEE335 pKa = 4.09SAEE338 pKa = 3.95ILLLQRR344 pKa = 11.84RR345 pKa = 11.84LRR347 pKa = 11.84ASMVLLVGDD356 pKa = 4.39VAQGKK361 pKa = 7.86ATTASSIEE369 pKa = 4.37YY370 pKa = 8.43LTLPVIYY377 pKa = 9.94RR378 pKa = 11.84SEE380 pKa = 3.83TTYY383 pKa = 11.49RR384 pKa = 11.84LGQEE388 pKa = 4.16TASLCSKK395 pKa = 10.02QGNRR399 pKa = 11.84MVSKK403 pKa = 10.59GGRR406 pKa = 11.84DD407 pKa = 3.25TVIITDD413 pKa = 3.63YY414 pKa = 11.28DD415 pKa = 3.95GEE417 pKa = 4.27TDD419 pKa = 3.39EE420 pKa = 4.62TEE422 pKa = 3.93KK423 pKa = 11.37NIAFTVDD430 pKa = 2.96TVRR433 pKa = 11.84DD434 pKa = 4.03VKK436 pKa = 11.31DD437 pKa = 3.59CGYY440 pKa = 10.99DD441 pKa = 3.1CALAIDD447 pKa = 4.19VQGKK451 pKa = 9.17EE452 pKa = 4.08FDD454 pKa = 3.71SVTLFLRR461 pKa = 11.84NEE463 pKa = 3.95DD464 pKa = 3.56RR465 pKa = 11.84KK466 pKa = 10.89ALADD470 pKa = 3.25KK471 pKa = 10.58HH472 pKa = 6.21LRR474 pKa = 11.84LVALSRR480 pKa = 11.84HH481 pKa = 5.11KK482 pKa = 10.93SKK484 pKa = 11.22LIIRR488 pKa = 11.84ADD490 pKa = 3.39AEE492 pKa = 3.77IRR494 pKa = 11.84QAFLTGDD501 pKa = 3.21IDD503 pKa = 4.76LSSKK507 pKa = 10.6ASNSHH512 pKa = 6.61RR513 pKa = 11.84YY514 pKa = 7.87SAKK517 pKa = 9.79PDD519 pKa = 3.68EE520 pKa = 4.29DD521 pKa = 4.02HH522 pKa = 7.41SWFKK526 pKa = 11.42AKK528 pKa = 10.47
MM1 pKa = 8.03DD2 pKa = 3.25MTKK5 pKa = 9.39TVEE8 pKa = 4.07EE9 pKa = 4.57KK10 pKa = 9.56KK11 pKa = 9.43TNGTDD16 pKa = 3.15SVKK19 pKa = 10.76GVFEE23 pKa = 4.28NSTIPKK29 pKa = 9.52VPTGQEE35 pKa = 3.63MGGDD39 pKa = 3.74GSSTSKK45 pKa = 10.8LKK47 pKa = 9.43EE48 pKa = 3.98TLKK51 pKa = 11.09VADD54 pKa = 3.99QTPLSVDD61 pKa = 2.98NGAKK65 pKa = 10.12SKK67 pKa = 10.84LDD69 pKa = 3.34SSDD72 pKa = 3.38RR73 pKa = 11.84QVPGVADD80 pKa = 3.68QTPLSVDD87 pKa = 2.98NGAKK91 pKa = 10.12SKK93 pKa = 10.84LDD95 pKa = 3.34SSDD98 pKa = 3.39RR99 pKa = 11.84QVPGPEE105 pKa = 4.03LKK107 pKa = 10.8PNVKK111 pKa = 9.88KK112 pKa = 10.8SKK114 pKa = 10.08KK115 pKa = 10.03KK116 pKa = 10.41RR117 pKa = 11.84IQKK120 pKa = 8.88PAQPSGPNDD129 pKa = 3.45LKK131 pKa = 11.36GGTKK135 pKa = 10.28GSSQVGEE142 pKa = 4.19NVSEE146 pKa = 4.21NYY148 pKa = 9.55TGISKK153 pKa = 10.04EE154 pKa = 3.99AAKK157 pKa = 10.32QKK159 pKa = 10.65QKK161 pKa = 9.58TPKK164 pKa = 8.97SVKK167 pKa = 9.0MQSNLADD174 pKa = 3.36KK175 pKa = 10.72FKK177 pKa = 11.41ANDD180 pKa = 3.36TRR182 pKa = 11.84RR183 pKa = 11.84SEE185 pKa = 4.85LINKK189 pKa = 7.42FQQFVHH195 pKa = 5.52EE196 pKa = 4.59TCLKK200 pKa = 10.69SDD202 pKa = 3.89FEE204 pKa = 4.61YY205 pKa = 10.4TGRR208 pKa = 11.84QYY210 pKa = 10.96FRR212 pKa = 11.84ARR214 pKa = 11.84SNFFEE219 pKa = 4.31MIKK222 pKa = 10.44LASLYY227 pKa = 10.48DD228 pKa = 3.56KK229 pKa = 10.82HH230 pKa = 8.26LKK232 pKa = 9.93EE233 pKa = 4.47CMARR237 pKa = 11.84ACTLEE242 pKa = 3.76RR243 pKa = 11.84EE244 pKa = 4.05RR245 pKa = 11.84LKK247 pKa = 10.99RR248 pKa = 11.84KK249 pKa = 9.57LLLVRR254 pKa = 11.84ALKK257 pKa = 10.01PAVDD261 pKa = 4.64FLTGIISGVPGSGKK275 pKa = 8.33STIVRR280 pKa = 11.84TLLKK284 pKa = 10.86GEE286 pKa = 4.48FPAVCALANPALMNDD301 pKa = 3.38YY302 pKa = 11.01SGIEE306 pKa = 4.06GVYY309 pKa = 10.94GLDD312 pKa = 4.2DD313 pKa = 4.69LLLSAVPITSDD324 pKa = 3.63LLIIDD329 pKa = 5.26EE330 pKa = 4.37YY331 pKa = 10.97TLAEE335 pKa = 4.09SAEE338 pKa = 3.95ILLLQRR344 pKa = 11.84RR345 pKa = 11.84LRR347 pKa = 11.84ASMVLLVGDD356 pKa = 4.39VAQGKK361 pKa = 7.86ATTASSIEE369 pKa = 4.37YY370 pKa = 8.43LTLPVIYY377 pKa = 9.94RR378 pKa = 11.84SEE380 pKa = 3.83TTYY383 pKa = 11.49RR384 pKa = 11.84LGQEE388 pKa = 4.16TASLCSKK395 pKa = 10.02QGNRR399 pKa = 11.84MVSKK403 pKa = 10.59GGRR406 pKa = 11.84DD407 pKa = 3.25TVIITDD413 pKa = 3.63YY414 pKa = 11.28DD415 pKa = 3.95GEE417 pKa = 4.27TDD419 pKa = 3.39EE420 pKa = 4.62TEE422 pKa = 3.93KK423 pKa = 11.37NIAFTVDD430 pKa = 2.96TVRR433 pKa = 11.84DD434 pKa = 4.03VKK436 pKa = 11.31DD437 pKa = 3.59CGYY440 pKa = 10.99DD441 pKa = 3.1CALAIDD447 pKa = 4.19VQGKK451 pKa = 9.17EE452 pKa = 4.08FDD454 pKa = 3.71SVTLFLRR461 pKa = 11.84NEE463 pKa = 3.95DD464 pKa = 3.56RR465 pKa = 11.84KK466 pKa = 10.89ALADD470 pKa = 3.25KK471 pKa = 10.58HH472 pKa = 6.21LRR474 pKa = 11.84LVALSRR480 pKa = 11.84HH481 pKa = 5.11KK482 pKa = 10.93SKK484 pKa = 11.22LIIRR488 pKa = 11.84ADD490 pKa = 3.39AEE492 pKa = 3.77IRR494 pKa = 11.84QAFLTGDD501 pKa = 3.21IDD503 pKa = 4.76LSSKK507 pKa = 10.6ASNSHH512 pKa = 6.61RR513 pKa = 11.84YY514 pKa = 7.87SAKK517 pKa = 9.79PDD519 pKa = 3.68EE520 pKa = 4.29DD521 pKa = 4.02HH522 pKa = 7.41SWFKK526 pKa = 11.42AKK528 pKa = 10.47
Molecular weight: 58.1 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3148 |
153 |
1139 |
449.7 |
50.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.417 ± 0.515 | 2.51 ± 0.489 |
6.163 ± 0.485 | 6.163 ± 0.298 |
4.416 ± 0.474 | 5.527 ± 0.537 |
2.097 ± 0.24 | 5.464 ± 0.448 |
6.766 ± 0.66 | 9.466 ± 0.447 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.128 ± 0.425 | 3.59 ± 0.238 |
4.479 ± 0.332 | 3.272 ± 0.326 |
5.813 ± 0.445 | 8.482 ± 0.68 |
5.273 ± 0.342 | 7.211 ± 0.648 |
0.985 ± 0.197 | 3.748 ± 0.344 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
