
Maribacter polysiphoniae
Taxonomy: cellular organisms; Bacteria; FCB group; Bacteroidetes/Chlorobi group; Bacteroidetes; Flavobacteriia; Flavobacteriales; Flavobacteriaceae; Maribacter
Average proteome isoelectric point is 6.44
Get precalculated fractions of proteins
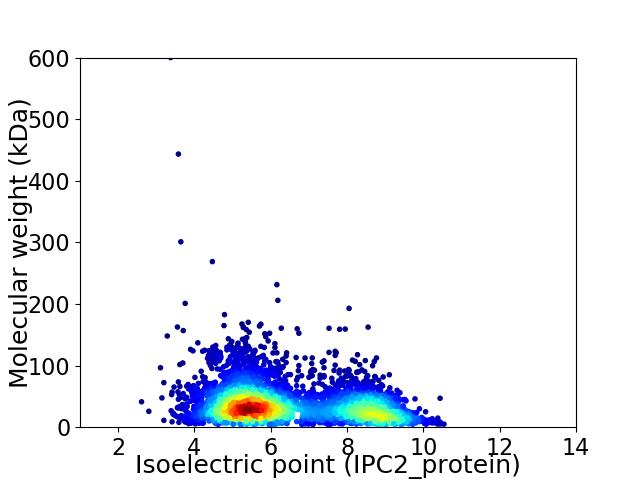
Virtual 2D-PAGE plot for 4431 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A316E4N3|A0A316E4N3_9FLAO 6-phosphogluconolactonase OS=Maribacter polysiphoniae OX=429344 GN=LX92_00103 PE=4 SV=1
MM1 pKa = 7.35KK2 pKa = 10.04PILRR6 pKa = 11.84TVMFGNSKK14 pKa = 9.81MISHH18 pKa = 7.24KK19 pKa = 10.99SFVCIIGVFIFFGFQNIFSQTTFTEE44 pKa = 4.47TAASFNLDD52 pKa = 2.6IGGNKK57 pKa = 10.14DD58 pKa = 4.18GGHH61 pKa = 6.55AWADD65 pKa = 3.46YY66 pKa = 11.53DD67 pKa = 4.47NDD69 pKa = 3.63GDD71 pKa = 4.78LDD73 pKa = 4.01VLVLEE78 pKa = 4.73NVNGSTRR85 pKa = 11.84SYY87 pKa = 11.5LMRR90 pKa = 11.84NNGNNTFTDD99 pKa = 3.26VRR101 pKa = 11.84AALITGMGNDD111 pKa = 3.23VAEE114 pKa = 4.62RR115 pKa = 11.84QAAWGDD121 pKa = 3.68LNNDD125 pKa = 3.09GRR127 pKa = 11.84PDD129 pKa = 3.69FMITSSGTSGGGGKK143 pKa = 9.77AALQIYY149 pKa = 7.94LQNTSGTFGDD159 pKa = 4.27GAGGNTPITVGRR171 pKa = 11.84GSFTININPINAEE184 pKa = 3.71GAGFFDD190 pKa = 5.29FEE192 pKa = 5.45GDD194 pKa = 3.18GDD196 pKa = 4.44LDD198 pKa = 4.12IFFDD202 pKa = 3.52SHH204 pKa = 7.59NYY206 pKa = 9.96GIEE209 pKa = 4.0LLRR212 pKa = 11.84NNYY215 pKa = 8.89IDD217 pKa = 3.91HH218 pKa = 5.76TTSTIVNPPAASLFTHH234 pKa = 6.22ITTGNGTGVVNFGLNQFATDD254 pKa = 3.52GDD256 pKa = 4.24YY257 pKa = 11.67GSAADD262 pKa = 4.13VNDD265 pKa = 5.14DD266 pKa = 3.27GWVDD270 pKa = 2.55IFMRR274 pKa = 11.84KK275 pKa = 8.61RR276 pKa = 11.84DD277 pKa = 3.74EE278 pKa = 4.48NDD280 pKa = 2.67FFLNQGGTFTNGADD294 pKa = 3.59LAQAEE299 pKa = 4.4NGNKK303 pKa = 9.74GGNGLWDD310 pKa = 4.35LDD312 pKa = 3.81NDD314 pKa = 4.14GDD316 pKa = 5.06LDD318 pKa = 4.86AVWTEE323 pKa = 3.75NGFTQIYY330 pKa = 10.08RR331 pKa = 11.84NDD333 pKa = 3.65AGTWTPLGAAVFPGLPQPSSSNSGASSARR362 pKa = 11.84IDD364 pKa = 3.59ALAGGDD370 pKa = 3.72IDD372 pKa = 4.66NDD374 pKa = 3.56GDD376 pKa = 3.96IDD378 pKa = 3.7ILLVGRR384 pKa = 11.84NRR386 pKa = 11.84SYY388 pKa = 11.62LYY390 pKa = 10.21INQLNSPTPAPGVVGSGTAMSFSLDD415 pKa = 3.37SQTFNSGRR423 pKa = 11.84DD424 pKa = 3.82GEE426 pKa = 4.7GTTMVDD432 pKa = 3.05VDD434 pKa = 5.18DD435 pKa = 6.66DD436 pKa = 4.8GDD438 pKa = 4.09LDD440 pKa = 3.71IYY442 pKa = 11.07MNINGNKK449 pKa = 9.46NKK451 pKa = 10.45LYY453 pKa = 10.8LNNLPAANRR462 pKa = 11.84KK463 pKa = 8.21NHH465 pKa = 6.3LLVDD469 pKa = 3.67VTEE472 pKa = 5.02DD473 pKa = 3.41RR474 pKa = 11.84DD475 pKa = 4.04ANGNTGGFPGRR486 pKa = 11.84VAIGTNVLIRR496 pKa = 11.84DD497 pKa = 3.78CAGNIVSGLRR507 pKa = 11.84QVNGVYY513 pKa = 10.51GHH515 pKa = 6.48GTQQPEE521 pKa = 4.14EE522 pKa = 4.14VHH524 pKa = 6.65FGLPLGEE531 pKa = 4.25GEE533 pKa = 4.25TYY535 pKa = 10.03IIEE538 pKa = 3.91VHH540 pKa = 5.88YY541 pKa = 10.53PNFNDD546 pKa = 3.58PVNGITRR553 pKa = 11.84LIATAIAQPSTIPGTNHH570 pKa = 6.21YY571 pKa = 11.2SLTTTNAEE579 pKa = 4.45LIEE582 pKa = 4.55NPNAPIANDD591 pKa = 3.67DD592 pKa = 4.34LKK594 pKa = 10.38TVAHH598 pKa = 6.75GNTVSVQISLFDD610 pKa = 3.98NDD612 pKa = 4.68SEE614 pKa = 4.52PDD616 pKa = 3.49GEE618 pKa = 4.4NFFIEE623 pKa = 5.18SVVQPAIGSVVIDD636 pKa = 4.44DD637 pKa = 4.42ADD639 pKa = 4.05AGLVTYY645 pKa = 8.31TYY647 pKa = 11.06SAATPFSGTTFDD659 pKa = 3.79YY660 pKa = 10.62TITDD664 pKa = 3.65ATSNLCPAQGKK675 pKa = 10.04SDD677 pKa = 3.69TATVTIFEE685 pKa = 4.94PCTDD689 pKa = 3.42SSGLDD694 pKa = 3.3TDD696 pKa = 4.59GDD698 pKa = 4.2GFNNVCDD705 pKa = 5.39LDD707 pKa = 4.79DD708 pKa = 5.79DD709 pKa = 4.42NDD711 pKa = 4.93GILDD715 pKa = 3.59VVEE718 pKa = 4.37RR719 pKa = 11.84PKK721 pKa = 10.61TVLWVLDD728 pKa = 3.69GTITPDD734 pKa = 3.22QQNVIDD740 pKa = 4.56KK741 pKa = 8.04LTNLGYY747 pKa = 8.91TLTLADD753 pKa = 5.56DD754 pKa = 4.77NDD756 pKa = 4.12SQDD759 pKa = 3.31ANNYY763 pKa = 8.63SVTYY767 pKa = 7.1VHH769 pKa = 7.07PSVSSGTAFANISNLATTTNGVITSEE795 pKa = 3.82NALFDD800 pKa = 4.15EE801 pKa = 5.2LFGTSGAVGNPTTNLINIINNTHH824 pKa = 7.7PITLSLPLGNLDD836 pKa = 3.3IGSGDD841 pKa = 3.64FYY843 pKa = 11.44VNNVVTGTKK852 pKa = 10.25LGQHH856 pKa = 7.14PDD858 pKa = 2.99GTANLIAWEE867 pKa = 4.24VGEE870 pKa = 5.52AMDD873 pKa = 4.45TGIAPGRR880 pKa = 11.84RR881 pKa = 11.84VAAPHH886 pKa = 6.19TSHH889 pKa = 7.94DD890 pKa = 3.81GGLNSAGEE898 pKa = 4.36DD899 pKa = 3.62LLVSAILWTWALDD912 pKa = 3.61TDD914 pKa = 4.78GDD916 pKa = 4.25GLYY919 pKa = 10.75DD920 pKa = 5.89DD921 pKa = 6.16LDD923 pKa = 5.1LDD925 pKa = 4.2SDD927 pKa = 4.4GDD929 pKa = 4.24GIPDD933 pKa = 3.41NVEE936 pKa = 3.69AQTTLTYY943 pKa = 9.85TPPNGDD949 pKa = 3.17SNAIYY954 pKa = 10.21AANNGVNSSYY964 pKa = 11.07LGGLLPVNTDD974 pKa = 2.95GADD977 pKa = 3.32NPDD980 pKa = 3.95YY981 pKa = 11.48LDD983 pKa = 3.58VDD985 pKa = 4.3SDD987 pKa = 4.37NEE989 pKa = 4.26GHH991 pKa = 7.35GDD993 pKa = 3.38TTEE996 pKa = 4.14AGIALAGVDD1005 pKa = 4.16TDD1007 pKa = 5.51NDD1009 pKa = 4.14GLDD1012 pKa = 4.2DD1013 pKa = 6.76GIDD1016 pKa = 3.72TDD1018 pKa = 3.97LTGYY1022 pKa = 10.42DD1023 pKa = 4.37DD1024 pKa = 4.75PGGTIDD1030 pKa = 6.08DD1031 pKa = 4.71PLNTPLTLLDD1041 pKa = 3.88TDD1043 pKa = 4.02NDD1045 pKa = 3.68ATTGGDD1051 pKa = 3.12VDD1053 pKa = 5.74FRR1055 pKa = 11.84DD1056 pKa = 4.3ALDD1059 pKa = 4.51DD1060 pKa = 5.0RR1061 pKa = 11.84PDD1063 pKa = 3.44NDD1065 pKa = 4.32LDD1067 pKa = 4.72GIVDD1071 pKa = 4.7AEE1073 pKa = 5.57DD1074 pKa = 4.48FDD1076 pKa = 6.6DD1077 pKa = 6.12DD1078 pKa = 4.63NDD1080 pKa = 4.96GILDD1084 pKa = 3.58IDD1086 pKa = 4.07EE1087 pKa = 4.84GCGNLVVNGNFEE1099 pKa = 4.22AQDD1102 pKa = 3.45FSDD1105 pKa = 4.58AVEE1108 pKa = 4.45FPNGFTEE1115 pKa = 4.91AGGTFIGATYY1125 pKa = 8.14NTNPLTGWSYY1135 pKa = 8.06TQNMDD1140 pKa = 2.67GWVGNQRR1147 pKa = 11.84MSWSSNDD1154 pKa = 2.97FAPAYY1159 pKa = 9.92KK1160 pKa = 9.95GGQYY1164 pKa = 10.08IDD1166 pKa = 4.33VIGNNAASGGSNNVLTQIINTEE1188 pKa = 3.89PGEE1191 pKa = 4.32TYY1193 pKa = 10.09TFSFFWGEE1201 pKa = 4.16DD1202 pKa = 3.02VGHH1205 pKa = 6.6RR1206 pKa = 11.84VGDD1209 pKa = 3.73PVILNVSVLDD1219 pKa = 3.83SGSNSLMNQTLNTTAFGIVDD1239 pKa = 4.48NIVGPQNWYY1248 pKa = 10.22YY1249 pKa = 11.01FEE1251 pKa = 4.04QTFIATTNTTTLRR1264 pKa = 11.84FAATPSGTANGTAIDD1279 pKa = 3.82FVYY1282 pKa = 9.35VTKK1285 pKa = 10.98NGLCQDD1291 pKa = 3.43TDD1293 pKa = 3.55GDD1295 pKa = 4.44GVIDD1299 pKa = 5.66AFDD1302 pKa = 4.98LDD1304 pKa = 3.98SDD1306 pKa = 3.86NDD1308 pKa = 4.58GIYY1311 pKa = 10.79DD1312 pKa = 3.63AVEE1315 pKa = 4.32AGHH1318 pKa = 6.74GAAHH1322 pKa = 6.4TNGVVNGAIGTDD1334 pKa = 3.88GVPDD1338 pKa = 4.22AVQSTPNNEE1347 pKa = 3.73MVNYY1351 pKa = 9.61IVEE1354 pKa = 4.87DD1355 pKa = 3.73SDD1357 pKa = 4.35SDD1359 pKa = 3.97GSIDD1363 pKa = 5.41AIEE1366 pKa = 4.74SDD1368 pKa = 5.19ADD1370 pKa = 3.93DD1371 pKa = 5.48DD1372 pKa = 4.32GCNDD1376 pKa = 3.23VDD1378 pKa = 3.76EE1379 pKa = 5.71AGYY1382 pKa = 9.31TDD1384 pKa = 3.94GNNDD1388 pKa = 3.33GLLGPNPITVDD1399 pKa = 3.3GNGLVISGTDD1409 pKa = 3.66GYY1411 pKa = 7.04TTPADD1416 pKa = 3.46ANTNSTYY1423 pKa = 11.16DD1424 pKa = 3.52FQEE1427 pKa = 4.4AGSPASIVGQPSNITICPGCTGEE1450 pKa = 4.7IKK1452 pKa = 9.5ITGTDD1457 pKa = 2.62IDD1459 pKa = 4.17TYY1461 pKa = 10.68QWQLFDD1467 pKa = 3.93GGSWMDD1473 pKa = 3.88LSDD1476 pKa = 3.32GGIYY1480 pKa = 10.26SGTATDD1486 pKa = 4.95SLIITNPTTSDD1497 pKa = 2.45TGNQYY1502 pKa = 10.58RR1503 pKa = 11.84VIVSNSAFVCSGDD1516 pKa = 3.22ISATGVLTVAAKK1528 pKa = 9.49TIITNRR1534 pKa = 11.84RR1535 pKa = 11.84ITYY1538 pKa = 9.63RR1539 pKa = 11.84VNKK1542 pKa = 9.67NN1543 pKa = 2.78
MM1 pKa = 7.35KK2 pKa = 10.04PILRR6 pKa = 11.84TVMFGNSKK14 pKa = 9.81MISHH18 pKa = 7.24KK19 pKa = 10.99SFVCIIGVFIFFGFQNIFSQTTFTEE44 pKa = 4.47TAASFNLDD52 pKa = 2.6IGGNKK57 pKa = 10.14DD58 pKa = 4.18GGHH61 pKa = 6.55AWADD65 pKa = 3.46YY66 pKa = 11.53DD67 pKa = 4.47NDD69 pKa = 3.63GDD71 pKa = 4.78LDD73 pKa = 4.01VLVLEE78 pKa = 4.73NVNGSTRR85 pKa = 11.84SYY87 pKa = 11.5LMRR90 pKa = 11.84NNGNNTFTDD99 pKa = 3.26VRR101 pKa = 11.84AALITGMGNDD111 pKa = 3.23VAEE114 pKa = 4.62RR115 pKa = 11.84QAAWGDD121 pKa = 3.68LNNDD125 pKa = 3.09GRR127 pKa = 11.84PDD129 pKa = 3.69FMITSSGTSGGGGKK143 pKa = 9.77AALQIYY149 pKa = 7.94LQNTSGTFGDD159 pKa = 4.27GAGGNTPITVGRR171 pKa = 11.84GSFTININPINAEE184 pKa = 3.71GAGFFDD190 pKa = 5.29FEE192 pKa = 5.45GDD194 pKa = 3.18GDD196 pKa = 4.44LDD198 pKa = 4.12IFFDD202 pKa = 3.52SHH204 pKa = 7.59NYY206 pKa = 9.96GIEE209 pKa = 4.0LLRR212 pKa = 11.84NNYY215 pKa = 8.89IDD217 pKa = 3.91HH218 pKa = 5.76TTSTIVNPPAASLFTHH234 pKa = 6.22ITTGNGTGVVNFGLNQFATDD254 pKa = 3.52GDD256 pKa = 4.24YY257 pKa = 11.67GSAADD262 pKa = 4.13VNDD265 pKa = 5.14DD266 pKa = 3.27GWVDD270 pKa = 2.55IFMRR274 pKa = 11.84KK275 pKa = 8.61RR276 pKa = 11.84DD277 pKa = 3.74EE278 pKa = 4.48NDD280 pKa = 2.67FFLNQGGTFTNGADD294 pKa = 3.59LAQAEE299 pKa = 4.4NGNKK303 pKa = 9.74GGNGLWDD310 pKa = 4.35LDD312 pKa = 3.81NDD314 pKa = 4.14GDD316 pKa = 5.06LDD318 pKa = 4.86AVWTEE323 pKa = 3.75NGFTQIYY330 pKa = 10.08RR331 pKa = 11.84NDD333 pKa = 3.65AGTWTPLGAAVFPGLPQPSSSNSGASSARR362 pKa = 11.84IDD364 pKa = 3.59ALAGGDD370 pKa = 3.72IDD372 pKa = 4.66NDD374 pKa = 3.56GDD376 pKa = 3.96IDD378 pKa = 3.7ILLVGRR384 pKa = 11.84NRR386 pKa = 11.84SYY388 pKa = 11.62LYY390 pKa = 10.21INQLNSPTPAPGVVGSGTAMSFSLDD415 pKa = 3.37SQTFNSGRR423 pKa = 11.84DD424 pKa = 3.82GEE426 pKa = 4.7GTTMVDD432 pKa = 3.05VDD434 pKa = 5.18DD435 pKa = 6.66DD436 pKa = 4.8GDD438 pKa = 4.09LDD440 pKa = 3.71IYY442 pKa = 11.07MNINGNKK449 pKa = 9.46NKK451 pKa = 10.45LYY453 pKa = 10.8LNNLPAANRR462 pKa = 11.84KK463 pKa = 8.21NHH465 pKa = 6.3LLVDD469 pKa = 3.67VTEE472 pKa = 5.02DD473 pKa = 3.41RR474 pKa = 11.84DD475 pKa = 4.04ANGNTGGFPGRR486 pKa = 11.84VAIGTNVLIRR496 pKa = 11.84DD497 pKa = 3.78CAGNIVSGLRR507 pKa = 11.84QVNGVYY513 pKa = 10.51GHH515 pKa = 6.48GTQQPEE521 pKa = 4.14EE522 pKa = 4.14VHH524 pKa = 6.65FGLPLGEE531 pKa = 4.25GEE533 pKa = 4.25TYY535 pKa = 10.03IIEE538 pKa = 3.91VHH540 pKa = 5.88YY541 pKa = 10.53PNFNDD546 pKa = 3.58PVNGITRR553 pKa = 11.84LIATAIAQPSTIPGTNHH570 pKa = 6.21YY571 pKa = 11.2SLTTTNAEE579 pKa = 4.45LIEE582 pKa = 4.55NPNAPIANDD591 pKa = 3.67DD592 pKa = 4.34LKK594 pKa = 10.38TVAHH598 pKa = 6.75GNTVSVQISLFDD610 pKa = 3.98NDD612 pKa = 4.68SEE614 pKa = 4.52PDD616 pKa = 3.49GEE618 pKa = 4.4NFFIEE623 pKa = 5.18SVVQPAIGSVVIDD636 pKa = 4.44DD637 pKa = 4.42ADD639 pKa = 4.05AGLVTYY645 pKa = 8.31TYY647 pKa = 11.06SAATPFSGTTFDD659 pKa = 3.79YY660 pKa = 10.62TITDD664 pKa = 3.65ATSNLCPAQGKK675 pKa = 10.04SDD677 pKa = 3.69TATVTIFEE685 pKa = 4.94PCTDD689 pKa = 3.42SSGLDD694 pKa = 3.3TDD696 pKa = 4.59GDD698 pKa = 4.2GFNNVCDD705 pKa = 5.39LDD707 pKa = 4.79DD708 pKa = 5.79DD709 pKa = 4.42NDD711 pKa = 4.93GILDD715 pKa = 3.59VVEE718 pKa = 4.37RR719 pKa = 11.84PKK721 pKa = 10.61TVLWVLDD728 pKa = 3.69GTITPDD734 pKa = 3.22QQNVIDD740 pKa = 4.56KK741 pKa = 8.04LTNLGYY747 pKa = 8.91TLTLADD753 pKa = 5.56DD754 pKa = 4.77NDD756 pKa = 4.12SQDD759 pKa = 3.31ANNYY763 pKa = 8.63SVTYY767 pKa = 7.1VHH769 pKa = 7.07PSVSSGTAFANISNLATTTNGVITSEE795 pKa = 3.82NALFDD800 pKa = 4.15EE801 pKa = 5.2LFGTSGAVGNPTTNLINIINNTHH824 pKa = 7.7PITLSLPLGNLDD836 pKa = 3.3IGSGDD841 pKa = 3.64FYY843 pKa = 11.44VNNVVTGTKK852 pKa = 10.25LGQHH856 pKa = 7.14PDD858 pKa = 2.99GTANLIAWEE867 pKa = 4.24VGEE870 pKa = 5.52AMDD873 pKa = 4.45TGIAPGRR880 pKa = 11.84RR881 pKa = 11.84VAAPHH886 pKa = 6.19TSHH889 pKa = 7.94DD890 pKa = 3.81GGLNSAGEE898 pKa = 4.36DD899 pKa = 3.62LLVSAILWTWALDD912 pKa = 3.61TDD914 pKa = 4.78GDD916 pKa = 4.25GLYY919 pKa = 10.75DD920 pKa = 5.89DD921 pKa = 6.16LDD923 pKa = 5.1LDD925 pKa = 4.2SDD927 pKa = 4.4GDD929 pKa = 4.24GIPDD933 pKa = 3.41NVEE936 pKa = 3.69AQTTLTYY943 pKa = 9.85TPPNGDD949 pKa = 3.17SNAIYY954 pKa = 10.21AANNGVNSSYY964 pKa = 11.07LGGLLPVNTDD974 pKa = 2.95GADD977 pKa = 3.32NPDD980 pKa = 3.95YY981 pKa = 11.48LDD983 pKa = 3.58VDD985 pKa = 4.3SDD987 pKa = 4.37NEE989 pKa = 4.26GHH991 pKa = 7.35GDD993 pKa = 3.38TTEE996 pKa = 4.14AGIALAGVDD1005 pKa = 4.16TDD1007 pKa = 5.51NDD1009 pKa = 4.14GLDD1012 pKa = 4.2DD1013 pKa = 6.76GIDD1016 pKa = 3.72TDD1018 pKa = 3.97LTGYY1022 pKa = 10.42DD1023 pKa = 4.37DD1024 pKa = 4.75PGGTIDD1030 pKa = 6.08DD1031 pKa = 4.71PLNTPLTLLDD1041 pKa = 3.88TDD1043 pKa = 4.02NDD1045 pKa = 3.68ATTGGDD1051 pKa = 3.12VDD1053 pKa = 5.74FRR1055 pKa = 11.84DD1056 pKa = 4.3ALDD1059 pKa = 4.51DD1060 pKa = 5.0RR1061 pKa = 11.84PDD1063 pKa = 3.44NDD1065 pKa = 4.32LDD1067 pKa = 4.72GIVDD1071 pKa = 4.7AEE1073 pKa = 5.57DD1074 pKa = 4.48FDD1076 pKa = 6.6DD1077 pKa = 6.12DD1078 pKa = 4.63NDD1080 pKa = 4.96GILDD1084 pKa = 3.58IDD1086 pKa = 4.07EE1087 pKa = 4.84GCGNLVVNGNFEE1099 pKa = 4.22AQDD1102 pKa = 3.45FSDD1105 pKa = 4.58AVEE1108 pKa = 4.45FPNGFTEE1115 pKa = 4.91AGGTFIGATYY1125 pKa = 8.14NTNPLTGWSYY1135 pKa = 8.06TQNMDD1140 pKa = 2.67GWVGNQRR1147 pKa = 11.84MSWSSNDD1154 pKa = 2.97FAPAYY1159 pKa = 9.92KK1160 pKa = 9.95GGQYY1164 pKa = 10.08IDD1166 pKa = 4.33VIGNNAASGGSNNVLTQIINTEE1188 pKa = 3.89PGEE1191 pKa = 4.32TYY1193 pKa = 10.09TFSFFWGEE1201 pKa = 4.16DD1202 pKa = 3.02VGHH1205 pKa = 6.6RR1206 pKa = 11.84VGDD1209 pKa = 3.73PVILNVSVLDD1219 pKa = 3.83SGSNSLMNQTLNTTAFGIVDD1239 pKa = 4.48NIVGPQNWYY1248 pKa = 10.22YY1249 pKa = 11.01FEE1251 pKa = 4.04QTFIATTNTTTLRR1264 pKa = 11.84FAATPSGTANGTAIDD1279 pKa = 3.82FVYY1282 pKa = 9.35VTKK1285 pKa = 10.98NGLCQDD1291 pKa = 3.43TDD1293 pKa = 3.55GDD1295 pKa = 4.44GVIDD1299 pKa = 5.66AFDD1302 pKa = 4.98LDD1304 pKa = 3.98SDD1306 pKa = 3.86NDD1308 pKa = 4.58GIYY1311 pKa = 10.79DD1312 pKa = 3.63AVEE1315 pKa = 4.32AGHH1318 pKa = 6.74GAAHH1322 pKa = 6.4TNGVVNGAIGTDD1334 pKa = 3.88GVPDD1338 pKa = 4.22AVQSTPNNEE1347 pKa = 3.73MVNYY1351 pKa = 9.61IVEE1354 pKa = 4.87DD1355 pKa = 3.73SDD1357 pKa = 4.35SDD1359 pKa = 3.97GSIDD1363 pKa = 5.41AIEE1366 pKa = 4.74SDD1368 pKa = 5.19ADD1370 pKa = 3.93DD1371 pKa = 5.48DD1372 pKa = 4.32GCNDD1376 pKa = 3.23VDD1378 pKa = 3.76EE1379 pKa = 5.71AGYY1382 pKa = 9.31TDD1384 pKa = 3.94GNNDD1388 pKa = 3.33GLLGPNPITVDD1399 pKa = 3.3GNGLVISGTDD1409 pKa = 3.66GYY1411 pKa = 7.04TTPADD1416 pKa = 3.46ANTNSTYY1423 pKa = 11.16DD1424 pKa = 3.52FQEE1427 pKa = 4.4AGSPASIVGQPSNITICPGCTGEE1450 pKa = 4.7IKK1452 pKa = 9.5ITGTDD1457 pKa = 2.62IDD1459 pKa = 4.17TYY1461 pKa = 10.68QWQLFDD1467 pKa = 3.93GGSWMDD1473 pKa = 3.88LSDD1476 pKa = 3.32GGIYY1480 pKa = 10.26SGTATDD1486 pKa = 4.95SLIITNPTTSDD1497 pKa = 2.45TGNQYY1502 pKa = 10.58RR1503 pKa = 11.84VIVSNSAFVCSGDD1516 pKa = 3.22ISATGVLTVAAKK1528 pKa = 9.49TIITNRR1534 pKa = 11.84RR1535 pKa = 11.84ITYY1538 pKa = 9.63RR1539 pKa = 11.84VNKK1542 pKa = 9.67NN1543 pKa = 2.78
Molecular weight: 162.67 kDa
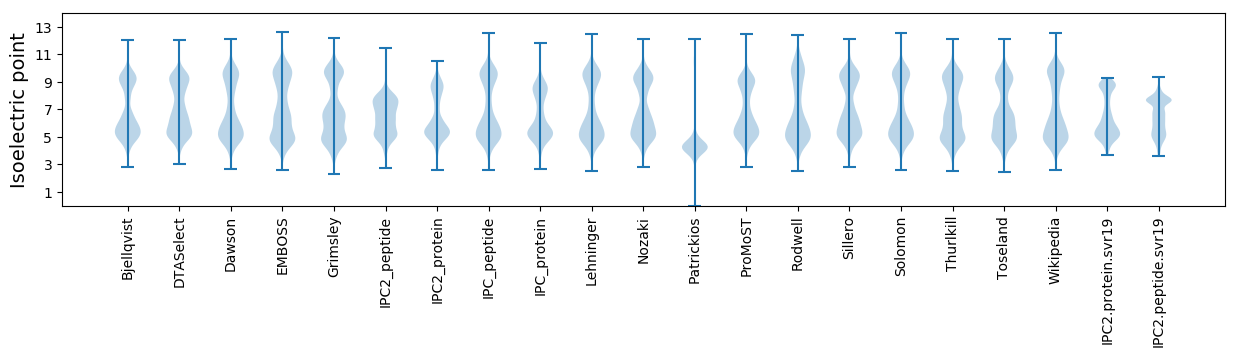
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A316E630|A0A316E630_9FLAO Copper chaperone CopZ OS=Maribacter polysiphoniae OX=429344 GN=LX92_01750 PE=4 SV=1
MM1 pKa = 7.21SRR3 pKa = 11.84EE4 pKa = 3.57RR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 10.21RR8 pKa = 11.84RR9 pKa = 11.84LEE11 pKa = 3.96EE12 pKa = 3.88KK13 pKa = 10.17KK14 pKa = 10.85GNSINRR20 pKa = 11.84KK21 pKa = 7.73VRR23 pKa = 11.84TRR25 pKa = 11.84KK26 pKa = 9.57EE27 pKa = 3.43KK28 pKa = 10.59GYY30 pKa = 10.43CLLVFVAMFLAVLTIAFLDD49 pKa = 4.49AILYY53 pKa = 9.21GFYY56 pKa = 11.18LLFNN60 pKa = 4.88
MM1 pKa = 7.21SRR3 pKa = 11.84EE4 pKa = 3.57RR5 pKa = 11.84RR6 pKa = 11.84KK7 pKa = 10.21RR8 pKa = 11.84RR9 pKa = 11.84LEE11 pKa = 3.96EE12 pKa = 3.88KK13 pKa = 10.17KK14 pKa = 10.85GNSINRR20 pKa = 11.84KK21 pKa = 7.73VRR23 pKa = 11.84TRR25 pKa = 11.84KK26 pKa = 9.57EE27 pKa = 3.43KK28 pKa = 10.59GYY30 pKa = 10.43CLLVFVAMFLAVLTIAFLDD49 pKa = 4.49AILYY53 pKa = 9.21GFYY56 pKa = 11.18LLFNN60 pKa = 4.88
Molecular weight: 7.19 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1523809 |
39 |
5722 |
343.9 |
38.73 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.416 ± 0.035 | 0.739 ± 0.011 |
5.891 ± 0.029 | 6.51 ± 0.036 |
5.039 ± 0.031 | 6.925 ± 0.035 |
1.889 ± 0.019 | 7.575 ± 0.038 |
7.439 ± 0.055 | 9.348 ± 0.044 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.416 ± 0.02 | 5.748 ± 0.035 |
3.615 ± 0.022 | 3.271 ± 0.017 |
3.593 ± 0.025 | 6.284 ± 0.03 |
5.755 ± 0.045 | 6.272 ± 0.027 |
1.172 ± 0.015 | 4.104 ± 0.023 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
