
Peanut clump virus (isolate 87/TGTA2) (PCV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Kitrinoviricota; Alsuviricetes; Martellivirales; Virgaviridae; Pecluvirus; Peanut clump virus
Average proteome isoelectric point is 7.05
Get precalculated fractions of proteins
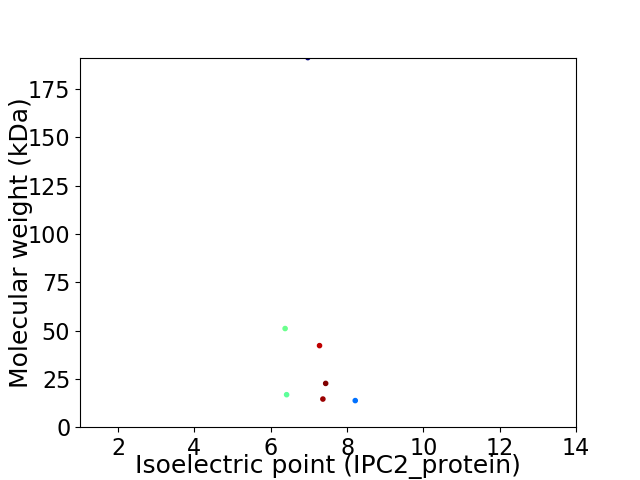
Virtual 2D-PAGE plot for 7 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions



Protein with the lowest isoelectric point:
>sp|Q08552|TGB3_PCV87 Movement protein TGB3 OS=Peanut clump virus (isolate 87/TGTA2) OX=652837 PE=3 SV=1
MM1 pKa = 6.91EE2 pKa = 4.07WRR4 pKa = 11.84RR5 pKa = 11.84AFSKK9 pKa = 10.69KK10 pKa = 10.1KK11 pKa = 10.65LEE13 pKa = 4.16EE14 pKa = 3.89KK15 pKa = 9.96VKK17 pKa = 11.15NMDD20 pKa = 3.12AKK22 pKa = 10.77QATDD26 pKa = 3.53YY27 pKa = 11.43LLEE30 pKa = 4.52KK31 pKa = 10.52VDD33 pKa = 3.66EE34 pKa = 4.15QRR36 pKa = 11.84NLEE39 pKa = 4.11NKK41 pKa = 9.77LDD43 pKa = 3.71KK44 pKa = 10.74RR45 pKa = 11.84LQNTRR50 pKa = 11.84KK51 pKa = 9.85KK52 pKa = 10.61NKK54 pKa = 9.47NKK56 pKa = 10.31EE57 pKa = 4.05KK58 pKa = 9.65TRR60 pKa = 11.84TWAEE64 pKa = 3.74KK65 pKa = 10.23YY66 pKa = 10.37PPVDD70 pKa = 3.33YY71 pKa = 10.94YY72 pKa = 11.63SPEE75 pKa = 3.77FVEE78 pKa = 5.85NFMKK82 pKa = 10.7DD83 pKa = 3.03MRR85 pKa = 11.84RR86 pKa = 11.84EE87 pKa = 3.8EE88 pKa = 4.2FEE90 pKa = 3.83KK91 pKa = 10.48SEE93 pKa = 3.93EE94 pKa = 4.15RR95 pKa = 11.84RR96 pKa = 11.84GHH98 pKa = 4.84KK99 pKa = 8.02QVRR102 pKa = 11.84LGSDD106 pKa = 3.15NFVGDD111 pKa = 4.55DD112 pKa = 3.66PLKK115 pKa = 10.68VLSEE119 pKa = 4.23EE120 pKa = 3.92ALKK123 pKa = 11.01AGFQHH128 pKa = 6.33TGKK131 pKa = 10.46VMKK134 pKa = 10.24RR135 pKa = 11.84FPADD139 pKa = 3.06VFEE142 pKa = 4.31KK143 pKa = 10.86SKK145 pKa = 10.99FIGMYY150 pKa = 9.95DD151 pKa = 3.1RR152 pKa = 11.84HH153 pKa = 5.91LTTLRR158 pKa = 11.84EE159 pKa = 3.95KK160 pKa = 10.7ACCKK164 pKa = 10.22KK165 pKa = 10.24EE166 pKa = 3.84RR167 pKa = 11.84NQIQSKK173 pKa = 10.16LIQLRR178 pKa = 11.84QLKK181 pKa = 9.55PSCDD185 pKa = 3.61FLAGTVSGVPGSGKK199 pKa = 8.62STLLKK204 pKa = 10.58NVQKK208 pKa = 10.71KK209 pKa = 10.06LKK211 pKa = 10.82NSVCLLANKK220 pKa = 8.26EE221 pKa = 4.19LKK223 pKa = 10.68GDD225 pKa = 3.81FAGVPSVFSVEE236 pKa = 4.04EE237 pKa = 4.03MLLSAVPSSFNVMLVDD253 pKa = 5.45EE254 pKa = 4.56YY255 pKa = 11.33TLTQSAEE262 pKa = 3.72ILLLQRR268 pKa = 11.84KK269 pKa = 8.82LGAKK273 pKa = 9.47IVVLFGDD280 pKa = 4.1RR281 pKa = 11.84EE282 pKa = 4.06QGNTNKK288 pKa = 8.89LTSPEE293 pKa = 4.0WLHH296 pKa = 6.05VPIVFSSDD304 pKa = 2.66SSHH307 pKa = 7.14RR308 pKa = 11.84FGPEE312 pKa = 3.2TAKK315 pKa = 10.67FCEE318 pKa = 4.21DD319 pKa = 2.9QGFSLEE325 pKa = 4.13GRR327 pKa = 11.84GGEE330 pKa = 4.08DD331 pKa = 3.63KK332 pKa = 10.58IVKK335 pKa = 9.85GDD337 pKa = 3.68YY338 pKa = 10.18EE339 pKa = 4.88GEE341 pKa = 4.42GEE343 pKa = 4.09DD344 pKa = 4.72TEE346 pKa = 5.19VNLCFTEE353 pKa = 4.21EE354 pKa = 4.01TKK356 pKa = 11.12ADD358 pKa = 3.73LAEE361 pKa = 4.27VQVEE365 pKa = 4.18AFLVSSVQGRR375 pKa = 11.84TFSSVSLFVRR385 pKa = 11.84EE386 pKa = 3.41NDD388 pKa = 3.37KK389 pKa = 11.1PVFSDD394 pKa = 2.82PHH396 pKa = 6.94LRR398 pKa = 11.84LVAITRR404 pKa = 11.84HH405 pKa = 5.75RR406 pKa = 11.84KK407 pKa = 9.08LLSIRR412 pKa = 11.84ADD414 pKa = 3.39PEE416 pKa = 4.05VWVSFMFATRR426 pKa = 11.84EE427 pKa = 4.1GEE429 pKa = 4.26EE430 pKa = 4.35VDD432 pKa = 3.35THH434 pKa = 7.01CYY436 pKa = 10.33GEE438 pKa = 3.99EE439 pKa = 3.93HH440 pKa = 6.94RR441 pKa = 11.84PDD443 pKa = 3.21EE444 pKa = 4.85AEE446 pKa = 3.76
MM1 pKa = 6.91EE2 pKa = 4.07WRR4 pKa = 11.84RR5 pKa = 11.84AFSKK9 pKa = 10.69KK10 pKa = 10.1KK11 pKa = 10.65LEE13 pKa = 4.16EE14 pKa = 3.89KK15 pKa = 9.96VKK17 pKa = 11.15NMDD20 pKa = 3.12AKK22 pKa = 10.77QATDD26 pKa = 3.53YY27 pKa = 11.43LLEE30 pKa = 4.52KK31 pKa = 10.52VDD33 pKa = 3.66EE34 pKa = 4.15QRR36 pKa = 11.84NLEE39 pKa = 4.11NKK41 pKa = 9.77LDD43 pKa = 3.71KK44 pKa = 10.74RR45 pKa = 11.84LQNTRR50 pKa = 11.84KK51 pKa = 9.85KK52 pKa = 10.61NKK54 pKa = 9.47NKK56 pKa = 10.31EE57 pKa = 4.05KK58 pKa = 9.65TRR60 pKa = 11.84TWAEE64 pKa = 3.74KK65 pKa = 10.23YY66 pKa = 10.37PPVDD70 pKa = 3.33YY71 pKa = 10.94YY72 pKa = 11.63SPEE75 pKa = 3.77FVEE78 pKa = 5.85NFMKK82 pKa = 10.7DD83 pKa = 3.03MRR85 pKa = 11.84RR86 pKa = 11.84EE87 pKa = 3.8EE88 pKa = 4.2FEE90 pKa = 3.83KK91 pKa = 10.48SEE93 pKa = 3.93EE94 pKa = 4.15RR95 pKa = 11.84RR96 pKa = 11.84GHH98 pKa = 4.84KK99 pKa = 8.02QVRR102 pKa = 11.84LGSDD106 pKa = 3.15NFVGDD111 pKa = 4.55DD112 pKa = 3.66PLKK115 pKa = 10.68VLSEE119 pKa = 4.23EE120 pKa = 3.92ALKK123 pKa = 11.01AGFQHH128 pKa = 6.33TGKK131 pKa = 10.46VMKK134 pKa = 10.24RR135 pKa = 11.84FPADD139 pKa = 3.06VFEE142 pKa = 4.31KK143 pKa = 10.86SKK145 pKa = 10.99FIGMYY150 pKa = 9.95DD151 pKa = 3.1RR152 pKa = 11.84HH153 pKa = 5.91LTTLRR158 pKa = 11.84EE159 pKa = 3.95KK160 pKa = 10.7ACCKK164 pKa = 10.22KK165 pKa = 10.24EE166 pKa = 3.84RR167 pKa = 11.84NQIQSKK173 pKa = 10.16LIQLRR178 pKa = 11.84QLKK181 pKa = 9.55PSCDD185 pKa = 3.61FLAGTVSGVPGSGKK199 pKa = 8.62STLLKK204 pKa = 10.58NVQKK208 pKa = 10.71KK209 pKa = 10.06LKK211 pKa = 10.82NSVCLLANKK220 pKa = 8.26EE221 pKa = 4.19LKK223 pKa = 10.68GDD225 pKa = 3.81FAGVPSVFSVEE236 pKa = 4.04EE237 pKa = 4.03MLLSAVPSSFNVMLVDD253 pKa = 5.45EE254 pKa = 4.56YY255 pKa = 11.33TLTQSAEE262 pKa = 3.72ILLLQRR268 pKa = 11.84KK269 pKa = 8.82LGAKK273 pKa = 9.47IVVLFGDD280 pKa = 4.1RR281 pKa = 11.84EE282 pKa = 4.06QGNTNKK288 pKa = 8.89LTSPEE293 pKa = 4.0WLHH296 pKa = 6.05VPIVFSSDD304 pKa = 2.66SSHH307 pKa = 7.14RR308 pKa = 11.84FGPEE312 pKa = 3.2TAKK315 pKa = 10.67FCEE318 pKa = 4.21DD319 pKa = 2.9QGFSLEE325 pKa = 4.13GRR327 pKa = 11.84GGEE330 pKa = 4.08DD331 pKa = 3.63KK332 pKa = 10.58IVKK335 pKa = 9.85GDD337 pKa = 3.68YY338 pKa = 10.18EE339 pKa = 4.88GEE341 pKa = 4.42GEE343 pKa = 4.09DD344 pKa = 4.72TEE346 pKa = 5.19VNLCFTEE353 pKa = 4.21EE354 pKa = 4.01TKK356 pKa = 11.12ADD358 pKa = 3.73LAEE361 pKa = 4.27VQVEE365 pKa = 4.18AFLVSSVQGRR375 pKa = 11.84TFSSVSLFVRR385 pKa = 11.84EE386 pKa = 3.41NDD388 pKa = 3.37KK389 pKa = 11.1PVFSDD394 pKa = 2.82PHH396 pKa = 6.94LRR398 pKa = 11.84LVAITRR404 pKa = 11.84HH405 pKa = 5.75RR406 pKa = 11.84KK407 pKa = 9.08LLSIRR412 pKa = 11.84ADD414 pKa = 3.39PEE416 pKa = 4.05VWVSFMFATRR426 pKa = 11.84EE427 pKa = 4.1GEE429 pKa = 4.26EE430 pKa = 4.35VDD432 pKa = 3.35THH434 pKa = 7.01CYY436 pKa = 10.33GEE438 pKa = 3.99EE439 pKa = 3.93HH440 pKa = 6.94RR441 pKa = 11.84PDD443 pKa = 3.21EE444 pKa = 4.85AEE446 pKa = 3.76
Molecular weight: 51.06 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q08316|P39_PCV87 Protein P39 OS=Peanut clump virus (isolate 87/TGTA2) OX=652837 PE=4 SV=1
MM1 pKa = 7.21VKK3 pKa = 9.07STVPTRR9 pKa = 11.84PNKK12 pKa = 9.03YY13 pKa = 7.94WPGVVAIGLVSLFIFLSVSNQKK35 pKa = 10.42HH36 pKa = 4.66STTSGDD42 pKa = 3.85NIHH45 pKa = 6.69KK46 pKa = 9.94FSNGGTYY53 pKa = 9.73RR54 pKa = 11.84DD55 pKa = 3.52GSKK58 pKa = 10.71CITYY62 pKa = 10.28NRR64 pKa = 11.84NSPLAYY70 pKa = 10.26NGSSSNNTLFWLCLLGLSMVWIAYY94 pKa = 9.43CGYY97 pKa = 10.69KK98 pKa = 10.11SLSGQWHH105 pKa = 6.19SCQHH109 pKa = 6.59DD110 pKa = 3.64KK111 pKa = 11.6NEE113 pKa = 4.19RR114 pKa = 11.84NFLFEE119 pKa = 4.32CFEE122 pKa = 4.15
MM1 pKa = 7.21VKK3 pKa = 9.07STVPTRR9 pKa = 11.84PNKK12 pKa = 9.03YY13 pKa = 7.94WPGVVAIGLVSLFIFLSVSNQKK35 pKa = 10.42HH36 pKa = 4.66STTSGDD42 pKa = 3.85NIHH45 pKa = 6.69KK46 pKa = 9.94FSNGGTYY53 pKa = 9.73RR54 pKa = 11.84DD55 pKa = 3.52GSKK58 pKa = 10.71CITYY62 pKa = 10.28NRR64 pKa = 11.84NSPLAYY70 pKa = 10.26NGSSSNNTLFWLCLLGLSMVWIAYY94 pKa = 9.43CGYY97 pKa = 10.69KK98 pKa = 10.11SLSGQWHH105 pKa = 6.19SCQHH109 pKa = 6.59DD110 pKa = 3.64KK111 pKa = 11.6NEE113 pKa = 4.19RR114 pKa = 11.84NFLFEE119 pKa = 4.32CFEE122 pKa = 4.15
Molecular weight: 13.77 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3091 |
122 |
1672 |
441.6 |
50.34 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.597 ± 0.36 | 2.297 ± 0.357 |
5.791 ± 0.319 | 7.409 ± 0.829 |
4.756 ± 0.215 | 5.532 ± 0.574 |
1.973 ± 0.236 | 4.109 ± 0.419 |
7.667 ± 0.646 | 9.803 ± 0.282 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.168 ± 0.249 | 4.012 ± 0.432 |
3.785 ± 0.237 | 3.429 ± 0.506 |
5.985 ± 0.591 | 7.894 ± 0.5 |
4.562 ± 0.37 | 8.153 ± 0.293 |
1.326 ± 0.21 | 3.72 ± 0.567 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
