
Pteropus associated gemycircularvirus 5
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Repensiviricetes; Geplafuvirales; Genomoviridae; Gemycircularvirus; Gemycircularvirus ptero5
Average proteome isoelectric point is 6.68
Get precalculated fractions of proteins
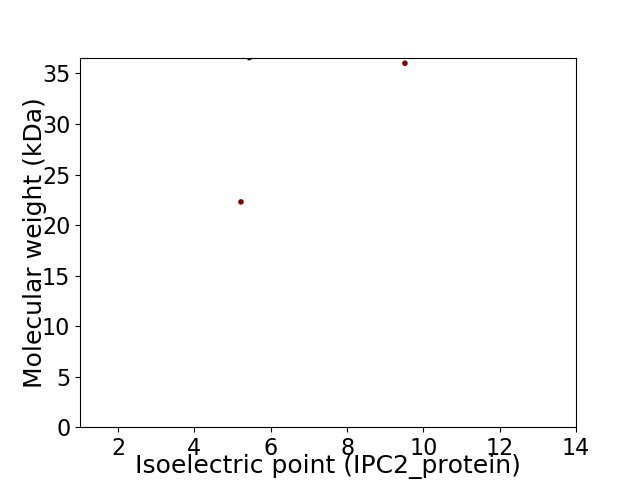
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A140CTN7|A0A140CTN7_9VIRU RepA OS=Pteropus associated gemycircularvirus 5 OX=1985399 PE=3 SV=1
MM1 pKa = 7.75PSSFHH6 pKa = 6.1LKK8 pKa = 8.75NRR10 pKa = 11.84RR11 pKa = 11.84YY12 pKa = 10.59VLFTYY17 pKa = 10.25SQAGPDD23 pKa = 3.36FDD25 pKa = 3.8YY26 pKa = 10.74WAVVDD31 pKa = 5.09LLGDD35 pKa = 3.78MGAEE39 pKa = 4.13CIIGRR44 pKa = 11.84EE45 pKa = 3.95VHH47 pKa = 6.75ADD49 pKa = 3.31GGIHH53 pKa = 5.77FHH55 pKa = 6.37VFTDD59 pKa = 4.01FGRR62 pKa = 11.84LFSTRR67 pKa = 11.84KK68 pKa = 9.62VRR70 pKa = 11.84VFDD73 pKa = 4.04VGGKK77 pKa = 9.29HH78 pKa = 6.54PNIKK82 pKa = 10.26PIGRR86 pKa = 11.84TPAQAYY92 pKa = 9.94DD93 pKa = 3.67YY94 pKa = 10.4AIKK97 pKa = 10.96DD98 pKa = 3.52GDD100 pKa = 4.04VVAGGLGRR108 pKa = 11.84PGGDD112 pKa = 4.46CDD114 pKa = 3.84WDD116 pKa = 3.87PDD118 pKa = 4.19NFWAAATHH126 pKa = 6.07TGSSEE131 pKa = 4.05EE132 pKa = 4.05FLHH135 pKa = 7.11FCDD138 pKa = 4.33QLAPRR143 pKa = 11.84DD144 pKa = 4.37FIRR147 pKa = 11.84GFTNFRR153 pKa = 11.84AYY155 pKa = 11.08SNWKK159 pKa = 8.82FNPGIPEE166 pKa = 3.7YY167 pKa = 10.13DD168 pKa = 3.53QPDD171 pKa = 3.75GATYY175 pKa = 7.79DD176 pKa = 3.43TSAAPGIDD184 pKa = 3.03EE185 pKa = 4.71WLSQSLIGTGKK196 pKa = 9.78SRR198 pKa = 11.84EE199 pKa = 3.97RR200 pKa = 3.54
MM1 pKa = 7.75PSSFHH6 pKa = 6.1LKK8 pKa = 8.75NRR10 pKa = 11.84RR11 pKa = 11.84YY12 pKa = 10.59VLFTYY17 pKa = 10.25SQAGPDD23 pKa = 3.36FDD25 pKa = 3.8YY26 pKa = 10.74WAVVDD31 pKa = 5.09LLGDD35 pKa = 3.78MGAEE39 pKa = 4.13CIIGRR44 pKa = 11.84EE45 pKa = 3.95VHH47 pKa = 6.75ADD49 pKa = 3.31GGIHH53 pKa = 5.77FHH55 pKa = 6.37VFTDD59 pKa = 4.01FGRR62 pKa = 11.84LFSTRR67 pKa = 11.84KK68 pKa = 9.62VRR70 pKa = 11.84VFDD73 pKa = 4.04VGGKK77 pKa = 9.29HH78 pKa = 6.54PNIKK82 pKa = 10.26PIGRR86 pKa = 11.84TPAQAYY92 pKa = 9.94DD93 pKa = 3.67YY94 pKa = 10.4AIKK97 pKa = 10.96DD98 pKa = 3.52GDD100 pKa = 4.04VVAGGLGRR108 pKa = 11.84PGGDD112 pKa = 4.46CDD114 pKa = 3.84WDD116 pKa = 3.87PDD118 pKa = 4.19NFWAAATHH126 pKa = 6.07TGSSEE131 pKa = 4.05EE132 pKa = 4.05FLHH135 pKa = 7.11FCDD138 pKa = 4.33QLAPRR143 pKa = 11.84DD144 pKa = 4.37FIRR147 pKa = 11.84GFTNFRR153 pKa = 11.84AYY155 pKa = 11.08SNWKK159 pKa = 8.82FNPGIPEE166 pKa = 3.7YY167 pKa = 10.13DD168 pKa = 3.53QPDD171 pKa = 3.75GATYY175 pKa = 7.79DD176 pKa = 3.43TSAAPGIDD184 pKa = 3.03EE185 pKa = 4.71WLSQSLIGTGKK196 pKa = 9.78SRR198 pKa = 11.84EE199 pKa = 3.97RR200 pKa = 3.54
Molecular weight: 22.29 kDa
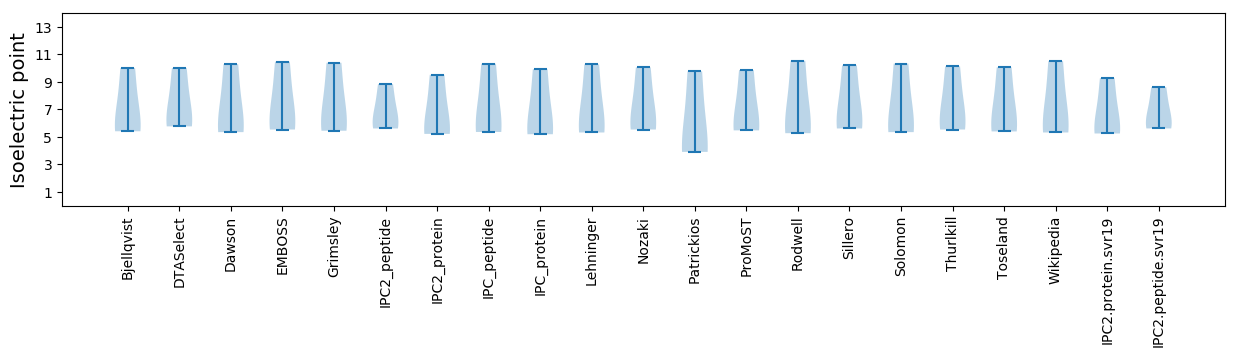
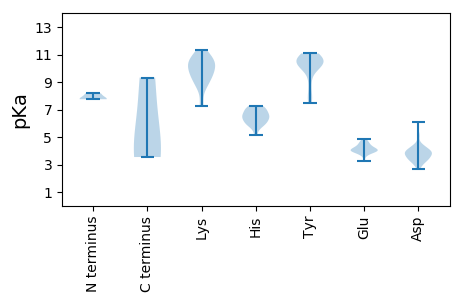
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A140CTN6|A0A140CTN6_9VIRU Replication-associated protein OS=Pteropus associated gemycircularvirus 5 OX=1985399 PE=3 SV=1
MM1 pKa = 8.18PYY3 pKa = 8.98RR4 pKa = 11.84TTRR7 pKa = 11.84RR8 pKa = 11.84GRR10 pKa = 11.84RR11 pKa = 11.84SFTRR15 pKa = 11.84KK16 pKa = 7.23VTPKK20 pKa = 10.36RR21 pKa = 11.84FGGTSRR27 pKa = 11.84RR28 pKa = 11.84TFPSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84FSGKK39 pKa = 9.47RR40 pKa = 11.84RR41 pKa = 11.84FTSKK45 pKa = 10.16RR46 pKa = 11.84MTRR49 pKa = 11.84KK50 pKa = 9.66KK51 pKa = 10.79VLDD54 pKa = 3.56YY55 pKa = 10.7TSKK58 pKa = 10.73KK59 pKa = 9.58KK60 pKa = 10.26QDD62 pKa = 2.94IMLQITNIDD71 pKa = 3.53AGGVTPGPLRR81 pKa = 11.84NGPSYY86 pKa = 10.96LQANKK91 pKa = 10.03TYY93 pKa = 10.14LIPFAATARR102 pKa = 11.84PAFGANLAPGNPADD116 pKa = 4.04RR117 pKa = 11.84AIRR120 pKa = 11.84TSSEE124 pKa = 3.84CYY126 pKa = 8.72MRR128 pKa = 11.84GIKK131 pKa = 9.74EE132 pKa = 4.33RR133 pKa = 11.84ITIRR137 pKa = 11.84TDD139 pKa = 4.11SGAPWQWRR147 pKa = 11.84RR148 pKa = 11.84ICIKK152 pKa = 9.46MRR154 pKa = 11.84GSKK157 pKa = 10.37FYY159 pKa = 10.55DD160 pKa = 3.38QEE162 pKa = 4.58SGTDD166 pKa = 3.31RR167 pKa = 11.84MLSYY171 pKa = 10.92LDD173 pKa = 4.13NADD176 pKa = 3.77IPQGMARR183 pKa = 11.84VATTWTQSDD192 pKa = 4.0SAWAVLCEE200 pKa = 3.99EE201 pKa = 4.83LFAGTRR207 pKa = 11.84GYY209 pKa = 10.9DD210 pKa = 2.97WSDD213 pKa = 3.17EE214 pKa = 3.95MLAPLDD220 pKa = 3.76KK221 pKa = 11.15NRR223 pKa = 11.84VSVAYY228 pKa = 10.07DD229 pKa = 3.21YY230 pKa = 7.48TTTIRR235 pKa = 11.84SGNDD239 pKa = 2.67SGVYY243 pKa = 8.07RR244 pKa = 11.84TVNRR248 pKa = 11.84WHH250 pKa = 7.12PMNKK254 pKa = 8.14TLYY257 pKa = 10.83YY258 pKa = 10.54DD259 pKa = 4.44DD260 pKa = 3.86QDD262 pKa = 4.04KK263 pKa = 11.33GDD265 pKa = 4.06RR266 pKa = 11.84LDD268 pKa = 3.35SRR270 pKa = 11.84RR271 pKa = 11.84WSMEE275 pKa = 3.24GNRR278 pKa = 11.84GMGDD282 pKa = 3.24YY283 pKa = 11.05YY284 pKa = 10.42IFDD287 pKa = 5.18FIKK290 pKa = 11.09GNGSPDD296 pKa = 3.38DD297 pKa = 3.77QLAIEE302 pKa = 4.54YY303 pKa = 10.75NSTLYY308 pKa = 9.56WHH310 pKa = 7.05EE311 pKa = 4.08KK312 pKa = 9.32
MM1 pKa = 8.18PYY3 pKa = 8.98RR4 pKa = 11.84TTRR7 pKa = 11.84RR8 pKa = 11.84GRR10 pKa = 11.84RR11 pKa = 11.84SFTRR15 pKa = 11.84KK16 pKa = 7.23VTPKK20 pKa = 10.36RR21 pKa = 11.84FGGTSRR27 pKa = 11.84RR28 pKa = 11.84TFPSRR33 pKa = 11.84RR34 pKa = 11.84RR35 pKa = 11.84FSGKK39 pKa = 9.47RR40 pKa = 11.84RR41 pKa = 11.84FTSKK45 pKa = 10.16RR46 pKa = 11.84MTRR49 pKa = 11.84KK50 pKa = 9.66KK51 pKa = 10.79VLDD54 pKa = 3.56YY55 pKa = 10.7TSKK58 pKa = 10.73KK59 pKa = 9.58KK60 pKa = 10.26QDD62 pKa = 2.94IMLQITNIDD71 pKa = 3.53AGGVTPGPLRR81 pKa = 11.84NGPSYY86 pKa = 10.96LQANKK91 pKa = 10.03TYY93 pKa = 10.14LIPFAATARR102 pKa = 11.84PAFGANLAPGNPADD116 pKa = 4.04RR117 pKa = 11.84AIRR120 pKa = 11.84TSSEE124 pKa = 3.84CYY126 pKa = 8.72MRR128 pKa = 11.84GIKK131 pKa = 9.74EE132 pKa = 4.33RR133 pKa = 11.84ITIRR137 pKa = 11.84TDD139 pKa = 4.11SGAPWQWRR147 pKa = 11.84RR148 pKa = 11.84ICIKK152 pKa = 9.46MRR154 pKa = 11.84GSKK157 pKa = 10.37FYY159 pKa = 10.55DD160 pKa = 3.38QEE162 pKa = 4.58SGTDD166 pKa = 3.31RR167 pKa = 11.84MLSYY171 pKa = 10.92LDD173 pKa = 4.13NADD176 pKa = 3.77IPQGMARR183 pKa = 11.84VATTWTQSDD192 pKa = 4.0SAWAVLCEE200 pKa = 3.99EE201 pKa = 4.83LFAGTRR207 pKa = 11.84GYY209 pKa = 10.9DD210 pKa = 2.97WSDD213 pKa = 3.17EE214 pKa = 3.95MLAPLDD220 pKa = 3.76KK221 pKa = 11.15NRR223 pKa = 11.84VSVAYY228 pKa = 10.07DD229 pKa = 3.21YY230 pKa = 7.48TTTIRR235 pKa = 11.84SGNDD239 pKa = 2.67SGVYY243 pKa = 8.07RR244 pKa = 11.84TVNRR248 pKa = 11.84WHH250 pKa = 7.12PMNKK254 pKa = 8.14TLYY257 pKa = 10.83YY258 pKa = 10.54DD259 pKa = 4.44DD260 pKa = 3.86QDD262 pKa = 4.04KK263 pKa = 11.33GDD265 pKa = 4.06RR266 pKa = 11.84LDD268 pKa = 3.35SRR270 pKa = 11.84RR271 pKa = 11.84WSMEE275 pKa = 3.24GNRR278 pKa = 11.84GMGDD282 pKa = 3.24YY283 pKa = 11.05YY284 pKa = 10.42IFDD287 pKa = 5.18FIKK290 pKa = 11.09GNGSPDD296 pKa = 3.38DD297 pKa = 3.77QLAIEE302 pKa = 4.54YY303 pKa = 10.75NSTLYY308 pKa = 9.56WHH310 pKa = 7.05EE311 pKa = 4.08KK312 pKa = 9.32
Molecular weight: 36.01 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
834 |
200 |
322 |
278.0 |
31.62 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
7.554 ± 0.531 | 1.559 ± 0.313 |
8.753 ± 0.528 | 3.597 ± 0.378 |
5.755 ± 1.026 | 9.592 ± 0.793 |
2.758 ± 0.988 | 5.036 ± 0.122 |
4.556 ± 0.564 | 5.635 ± 0.412 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.918 ± 0.734 | 3.237 ± 0.441 |
5.156 ± 0.275 | 2.398 ± 0.265 |
8.393 ± 1.584 | 5.995 ± 0.641 |
6.235 ± 1.108 | 4.197 ± 0.599 |
3.237 ± 0.516 | 4.436 ± 0.467 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
