
Melbournevirus
Taxonomy: Viruses; Varidnaviria; Bamfordvirae; Nucleocytoviricota; Megaviricetes; Pimascovirales; Marseilleviridae; Marseillevirus; unclassified Marseillevirus
Average proteome isoelectric point is 6.96
Get precalculated fractions of proteins
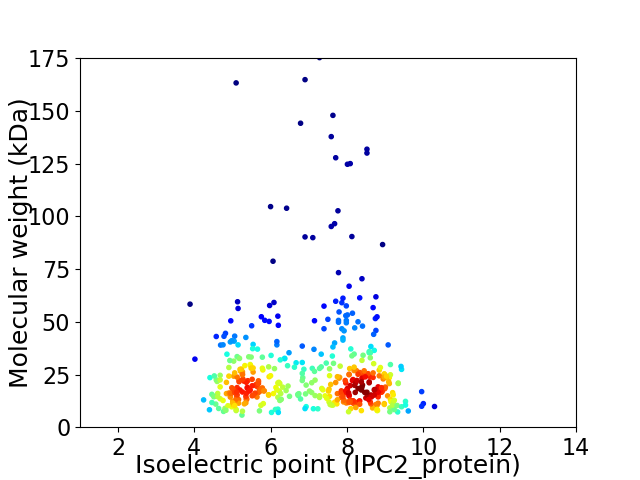
Virtual 2D-PAGE plot for 448 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
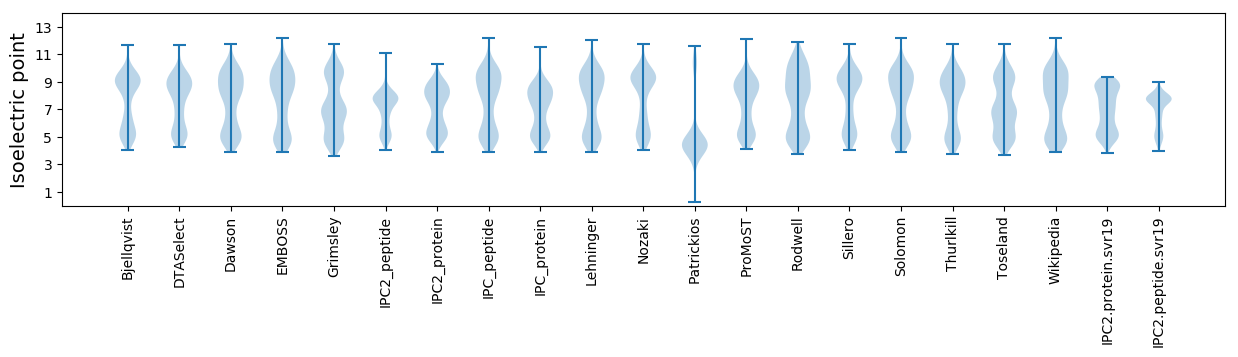
* You can choose from 21 different methods for calculating isoelectric point
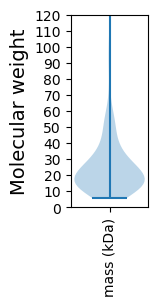
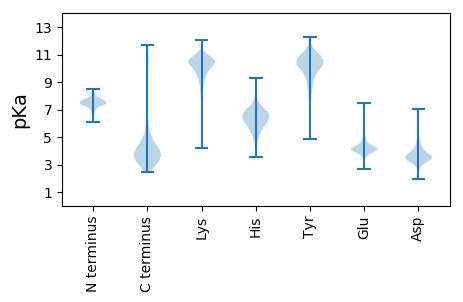
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A097I1F7|A0A097I1F7_9VIRU Uncharacterized protein OS=Melbournevirus OX=1560514 GN=MEL_044 PE=4 SV=1
MM1 pKa = 7.16SLVRR5 pKa = 11.84GFFKK9 pKa = 10.57DD10 pKa = 3.24IPIVGSYY17 pKa = 10.72SVLPPVLPRR26 pKa = 11.84GSLVFLTTDD35 pKa = 2.98QNLYY39 pKa = 10.15ISNGQIWAVAGGDD52 pKa = 3.84VGPLAAQVAQNTADD66 pKa = 3.38IATLQTDD73 pKa = 3.63VTTLDD78 pKa = 3.56GQVATNTSNITALQGQVATNTSDD101 pKa = 2.77IGTLQGQVATNTTDD115 pKa = 2.52ITTLQGQVATNTSNITTLQGQVATNTSNITTLQGQVATNTSDD157 pKa = 2.76ITSNTNSINNIIVSQSQGFTIADD180 pKa = 3.64SAQLRR185 pKa = 11.84TFTNPITRR193 pKa = 11.84IDD195 pKa = 3.71VSSTSPTVIYY205 pKa = 10.0TLGTPLDD212 pKa = 3.85GSFNGVYY219 pKa = 10.54LIQWNILAHH228 pKa = 5.22YY229 pKa = 7.21QTANVFYY236 pKa = 10.62AVGQSSVLSNGIGALVSVFGTSTNQTNSSGSSVITVTADD275 pKa = 2.85INLNNYY281 pKa = 9.3RR282 pKa = 11.84LIVTSDD288 pKa = 3.3SAVTTTYY295 pKa = 10.17SGYY298 pKa = 10.92VIVTRR303 pKa = 11.84ILTISPP309 pKa = 3.78
MM1 pKa = 7.16SLVRR5 pKa = 11.84GFFKK9 pKa = 10.57DD10 pKa = 3.24IPIVGSYY17 pKa = 10.72SVLPPVLPRR26 pKa = 11.84GSLVFLTTDD35 pKa = 2.98QNLYY39 pKa = 10.15ISNGQIWAVAGGDD52 pKa = 3.84VGPLAAQVAQNTADD66 pKa = 3.38IATLQTDD73 pKa = 3.63VTTLDD78 pKa = 3.56GQVATNTSNITALQGQVATNTSDD101 pKa = 2.77IGTLQGQVATNTTDD115 pKa = 2.52ITTLQGQVATNTSNITTLQGQVATNTSNITTLQGQVATNTSDD157 pKa = 2.76ITSNTNSINNIIVSQSQGFTIADD180 pKa = 3.64SAQLRR185 pKa = 11.84TFTNPITRR193 pKa = 11.84IDD195 pKa = 3.71VSSTSPTVIYY205 pKa = 10.0TLGTPLDD212 pKa = 3.85GSFNGVYY219 pKa = 10.54LIQWNILAHH228 pKa = 5.22YY229 pKa = 7.21QTANVFYY236 pKa = 10.62AVGQSSVLSNGIGALVSVFGTSTNQTNSSGSSVITVTADD275 pKa = 2.85INLNNYY281 pKa = 9.3RR282 pKa = 11.84LIVTSDD288 pKa = 3.3SAVTTTYY295 pKa = 10.17SGYY298 pKa = 10.92VIVTRR303 pKa = 11.84ILTISPP309 pKa = 3.78
Molecular weight: 32.29 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A097I1M7|A0A097I1M7_9VIRU Uncharacterized protein OS=Melbournevirus OX=1560514 GN=MEL_128 PE=4 SV=1
MM1 pKa = 7.78GYY3 pKa = 10.85LFGRR7 pKa = 11.84SWKK10 pKa = 9.63QKK12 pKa = 9.72RR13 pKa = 11.84IQNGKK18 pKa = 9.08SLGYY22 pKa = 8.83IQKK25 pKa = 9.8SRR27 pKa = 11.84SRR29 pKa = 11.84TTLPVLNSLLFLCFSQNISSPGKK52 pKa = 9.38EE53 pKa = 3.69RR54 pKa = 11.84PKK56 pKa = 9.83KK57 pKa = 8.39TRR59 pKa = 11.84KK60 pKa = 9.37LLYY63 pKa = 10.12GNVRR67 pKa = 11.84KK68 pKa = 10.0KK69 pKa = 11.07LPDD72 pKa = 3.31AALYY76 pKa = 10.0FKK78 pKa = 11.25GPFYY82 pKa = 11.0FLLFLRR88 pKa = 11.84FSFFSSFVDD97 pKa = 3.55LFFVYY102 pKa = 10.3SVPLPVPLCHH112 pKa = 6.89SFFIFFSVCSLSFRR126 pKa = 11.84SRR128 pKa = 11.84LCFALFHH135 pKa = 6.62IGCRR139 pKa = 11.84WKK141 pKa = 9.59TVTT144 pKa = 4.72
MM1 pKa = 7.78GYY3 pKa = 10.85LFGRR7 pKa = 11.84SWKK10 pKa = 9.63QKK12 pKa = 9.72RR13 pKa = 11.84IQNGKK18 pKa = 9.08SLGYY22 pKa = 8.83IQKK25 pKa = 9.8SRR27 pKa = 11.84SRR29 pKa = 11.84TTLPVLNSLLFLCFSQNISSPGKK52 pKa = 9.38EE53 pKa = 3.69RR54 pKa = 11.84PKK56 pKa = 9.83KK57 pKa = 8.39TRR59 pKa = 11.84KK60 pKa = 9.37LLYY63 pKa = 10.12GNVRR67 pKa = 11.84KK68 pKa = 10.0KK69 pKa = 11.07LPDD72 pKa = 3.31AALYY76 pKa = 10.0FKK78 pKa = 11.25GPFYY82 pKa = 11.0FLLFLRR88 pKa = 11.84FSFFSSFVDD97 pKa = 3.55LFFVYY102 pKa = 10.3SVPLPVPLCHH112 pKa = 6.89SFFIFFSVCSLSFRR126 pKa = 11.84SRR128 pKa = 11.84LCFALFHH135 pKa = 6.62IGCRR139 pKa = 11.84WKK141 pKa = 9.59TVTT144 pKa = 4.72
Molecular weight: 16.87 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
110121 |
50 |
1537 |
245.8 |
28.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.144 ± 0.123 | 2.532 ± 0.094 |
4.6 ± 0.081 | 8.86 ± 0.159 |
5.787 ± 0.104 | 6.277 ± 0.124 |
1.84 ± 0.06 | 4.834 ± 0.082 |
8.7 ± 0.184 | 8.898 ± 0.114 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.022 ± 0.046 | 3.832 ± 0.087 |
3.998 ± 0.101 | 3.341 ± 0.078 |
5.717 ± 0.102 | 7.772 ± 0.147 |
4.925 ± 0.129 | 6.338 ± 0.099 |
1.569 ± 0.046 | 3.012 ± 0.068 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
