
Bogoriella caseilytica
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Micrococcales; Bogoriellaceae; Bogoriella
Average proteome isoelectric point is 5.98
Get precalculated fractions of proteins
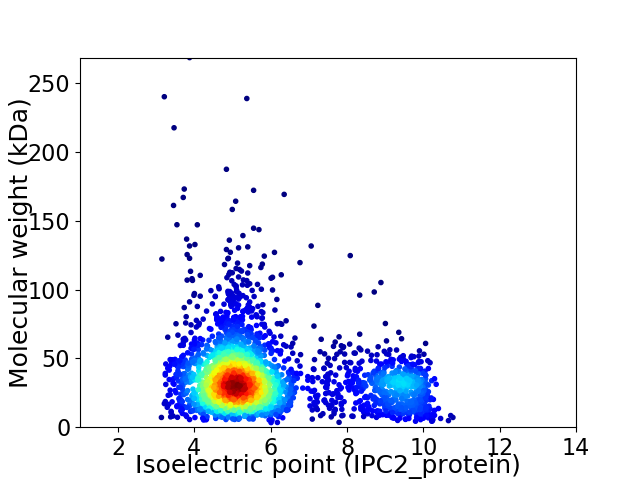
Virtual 2D-PAGE plot for 2844 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
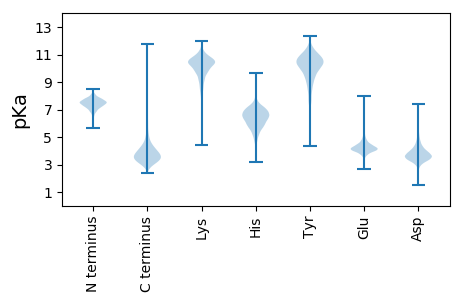
Protein with the lowest isoelectric point:
>tr|A0A3N2BEL8|A0A3N2BEL8_9MICO Enterochelin esterase family protein OS=Bogoriella caseilytica OX=56055 GN=EDD31_2091 PE=4 SV=1
MM1 pKa = 7.44AVLIGVLGSMAMAVPVAADD20 pKa = 3.49TALEE24 pKa = 3.96PAIEE28 pKa = 4.75ADD30 pKa = 4.48SADD33 pKa = 3.74ADD35 pKa = 3.8ADD37 pKa = 3.74ADD39 pKa = 3.97ADD41 pKa = 3.74ADD43 pKa = 3.97AAADD47 pKa = 3.61AAAEE51 pKa = 4.12ADD53 pKa = 3.83STEE56 pKa = 4.02AAAEE60 pKa = 4.05EE61 pKa = 4.21DD62 pKa = 4.37TEE64 pKa = 4.5TLSDD68 pKa = 3.62TADD71 pKa = 3.36EE72 pKa = 4.54GTPATSSDD80 pKa = 3.91DD81 pKa = 3.58VSTLAEE87 pKa = 3.87VSPFAEE93 pKa = 4.35PSPVYY98 pKa = 10.21EE99 pKa = 4.01ITGVWLEE106 pKa = 4.04QPEE109 pKa = 4.55VTSRR113 pKa = 11.84GNPVVADD120 pKa = 3.15WRR122 pKa = 11.84INVNDD127 pKa = 3.99GNNPPSNEE135 pKa = 3.78LVDD138 pKa = 4.2NVVATFDD145 pKa = 3.19VGNAVFQTIPDD156 pKa = 3.79LCLTTGVQPASSISADD172 pKa = 3.59GSLLTCNFGTVALGRR187 pKa = 11.84SIALQTPVITEE198 pKa = 4.28GVTGTNVSLSGTSPAGEE215 pKa = 4.11TVEE218 pKa = 5.3LPLIPIQNDD227 pKa = 2.77FRR229 pKa = 11.84MDD231 pKa = 3.14MHH233 pKa = 7.16FGQNTNQRR241 pKa = 11.84EE242 pKa = 4.16WDD244 pKa = 3.79SLSNPTNVNLDD255 pKa = 3.85VQWSLRR261 pKa = 11.84LGKK264 pKa = 10.77GSDD267 pKa = 3.72PGPANQTFRR276 pKa = 11.84LRR278 pKa = 11.84IAPIFPAGSTVEE290 pKa = 4.34VGTHH294 pKa = 6.4IYY296 pKa = 9.35TGVVGCSPFDD306 pKa = 3.75LSNAPGHH313 pKa = 6.81PYY315 pKa = 10.46SSNPPHH321 pKa = 6.65PRR323 pKa = 11.84HH324 pKa = 5.89TNFVDD329 pKa = 3.41SCTLTAVPGEE339 pKa = 4.12PGVFDD344 pKa = 4.03LTLEE348 pKa = 4.64GINYY352 pKa = 10.0SLVNAPTNDD361 pKa = 3.2AGTANSPLPADD372 pKa = 3.55WDD374 pKa = 4.28YY375 pKa = 11.82VASGSLWFQVNAGSNGSLALTMDD398 pKa = 4.43PVTYY402 pKa = 9.18TATTGQTSEE411 pKa = 5.0DD412 pKa = 3.89LGTNNQTTKK421 pKa = 11.17AFTLPGAWSASFRR434 pKa = 11.84RR435 pKa = 11.84SATGSGGTFWDD446 pKa = 3.49DD447 pKa = 3.37SYY449 pKa = 11.78RR450 pKa = 11.84VTPGVTLRR458 pKa = 11.84NEE460 pKa = 3.75ARR462 pKa = 11.84NGYY465 pKa = 10.06SGDD468 pKa = 3.59RR469 pKa = 11.84NVAPTAQYY477 pKa = 10.82GSCLALDD484 pKa = 3.66TRR486 pKa = 11.84YY487 pKa = 10.88VIVDD491 pKa = 3.56RR492 pKa = 11.84DD493 pKa = 3.23FDD495 pKa = 4.23LAVATGFLARR505 pKa = 11.84NDD507 pKa = 3.62WPASAVLEE515 pKa = 4.44YY516 pKa = 10.92YY517 pKa = 10.88VGGSTLLDD525 pKa = 3.87PATGNTAYY533 pKa = 10.87NPDD536 pKa = 3.55QFEE539 pKa = 4.54CDD541 pKa = 3.66SDD543 pKa = 3.84PGGWTTEE550 pKa = 4.05LPADD554 pKa = 4.38LSTIRR559 pKa = 11.84AARR562 pKa = 11.84VTYY565 pKa = 10.27DD566 pKa = 3.48FSSFDD571 pKa = 3.33EE572 pKa = 4.35VGRR575 pKa = 11.84EE576 pKa = 4.09FIDD579 pKa = 3.65LVGWTTVRR587 pKa = 11.84EE588 pKa = 4.05DD589 pKa = 3.49VQPGQDD595 pKa = 2.07IWTFGSRR602 pKa = 11.84LRR604 pKa = 11.84NGGWFGPAQEE614 pKa = 4.57EE615 pKa = 5.39VITPTPDD622 pKa = 2.52ARR624 pKa = 11.84YY625 pKa = 8.07PHH627 pKa = 6.65TDD629 pKa = 2.44GRR631 pKa = 11.84RR632 pKa = 11.84DD633 pKa = 3.2IARR636 pKa = 11.84VISANPAIQKK646 pKa = 9.26SAEE649 pKa = 3.98RR650 pKa = 11.84SVVVPGVPTVFTLRR664 pKa = 11.84YY665 pKa = 8.55SANTGAGIPEE675 pKa = 4.35SVDD678 pKa = 2.96GYY680 pKa = 9.1TVVDD684 pKa = 3.81TLPPLVTYY692 pKa = 9.71EE693 pKa = 4.28VGSSDD698 pKa = 4.18PEE700 pKa = 4.05PTVTTNANNQQVLTWTFDD718 pKa = 3.57NVPTNVQNVLTYY730 pKa = 10.73AAAADD735 pKa = 3.9SSVAPGTPLVNTATASLGGTTSSPARR761 pKa = 11.84ATVTTANNGFTTILKK776 pKa = 9.07TSDD779 pKa = 3.1VEE781 pKa = 4.59FIPNRR786 pKa = 11.84LGDD789 pKa = 4.08GVGSGSWTVTIEE801 pKa = 4.03SRR803 pKa = 11.84DD804 pKa = 3.85PQSQAWTDD812 pKa = 3.68TIDD815 pKa = 3.21ILPYY819 pKa = 10.53NGDD822 pKa = 3.13GRR824 pKa = 11.84GTSFSGTYY832 pKa = 9.37TLGDD836 pKa = 3.27VVLFDD841 pKa = 5.17GGTLYY846 pKa = 9.82YY847 pKa = 10.43TDD849 pKa = 5.36ADD851 pKa = 4.23PATLSDD857 pKa = 4.56DD858 pKa = 4.08PAHH861 pKa = 6.79EE862 pKa = 4.96SNGAPNAPSALWSTEE877 pKa = 3.66RR878 pKa = 11.84PEE880 pKa = 4.12NPTAIRR886 pKa = 11.84VIGGTLATGGSFSFQVPITTDD907 pKa = 3.02GAEE910 pKa = 4.15AQDD913 pKa = 3.61VFVNRR918 pKa = 11.84AQARR922 pKa = 11.84AEE924 pKa = 3.91HH925 pKa = 6.5TEE927 pKa = 3.72LVMRR931 pKa = 11.84TSAALVVTDD940 pKa = 3.55YY941 pKa = 11.32LVEE944 pKa = 4.24KK945 pKa = 10.22SANPEE950 pKa = 3.7SGSTVRR956 pKa = 11.84PGDD959 pKa = 3.58VVEE962 pKa = 4.26YY963 pKa = 9.29TVTVSQMGDD972 pKa = 3.17VPAGAWFSDD981 pKa = 3.68DD982 pKa = 3.71MSDD985 pKa = 4.18VLDD988 pKa = 4.42DD989 pKa = 4.02ADD991 pKa = 3.99YY992 pKa = 11.51NGDD995 pKa = 3.17VTASHH1000 pKa = 6.79GEE1002 pKa = 3.97VLSFEE1007 pKa = 4.28NGILEE1012 pKa = 4.37WEE1014 pKa = 4.6GVVPVGEE1021 pKa = 4.35IATITYY1027 pKa = 10.29SVTVKK1032 pKa = 10.44AAEE1035 pKa = 4.09EE1036 pKa = 3.85LRR1038 pKa = 11.84EE1039 pKa = 4.13GDD1041 pKa = 4.7SNTFLNNQVTSPGCQGACSTEE1062 pKa = 4.02HH1063 pKa = 6.03LVGYY1067 pKa = 9.57YY1068 pKa = 9.96SVSKK1072 pKa = 9.09TAEE1075 pKa = 4.03PAPGEE1080 pKa = 4.19TVRR1083 pKa = 11.84IGDD1086 pKa = 3.87VITYY1090 pKa = 8.78HH1091 pKa = 5.79VQVTQHH1097 pKa = 6.42GEE1099 pKa = 4.1GAVDD1103 pKa = 3.2NAFFIDD1109 pKa = 4.52DD1110 pKa = 4.15LSEE1113 pKa = 4.15VLDD1116 pKa = 3.96DD1117 pKa = 6.57AIWNDD1122 pKa = 3.64DD1123 pKa = 3.32AAATDD1128 pKa = 4.39GEE1130 pKa = 4.57LSFDD1134 pKa = 3.88EE1135 pKa = 6.13PILSWSAPLEE1145 pKa = 4.32VGDD1148 pKa = 4.39VVDD1151 pKa = 3.37VTYY1154 pKa = 11.08SVTVTGEE1161 pKa = 3.49GDD1163 pKa = 3.5YY1164 pKa = 11.05EE1165 pKa = 4.1LRR1167 pKa = 11.84NVVSSVCAEE1176 pKa = 4.62DD1177 pKa = 6.06DD1178 pKa = 4.05PDD1180 pKa = 4.21CLEE1183 pKa = 5.05AICVPAADD1191 pKa = 4.8EE1192 pKa = 5.12NPDD1195 pKa = 3.53CTTEE1199 pKa = 4.58HH1200 pKa = 6.59IVGDD1204 pKa = 3.86YY1205 pKa = 10.63QVTKK1209 pKa = 10.9VSDD1212 pKa = 4.07PEE1214 pKa = 4.14SGSAVEE1220 pKa = 4.34VGDD1223 pKa = 4.0VIDD1226 pKa = 3.76YY1227 pKa = 7.79TVTVSHH1233 pKa = 7.04IGLADD1238 pKa = 3.56VEE1240 pKa = 4.41ASFVDD1245 pKa = 3.97DD1246 pKa = 5.32LSDD1249 pKa = 4.22VLDD1252 pKa = 4.09DD1253 pKa = 6.15AIWNDD1258 pKa = 4.05DD1259 pKa = 3.28VSASAGEE1266 pKa = 4.04FSFEE1270 pKa = 4.67GEE1272 pKa = 4.21TLSWWGALSEE1282 pKa = 4.31GDD1284 pKa = 4.0VVTVTYY1290 pKa = 10.65SVTVTAEE1297 pKa = 3.63GDD1299 pKa = 3.38RR1300 pKa = 11.84YY1301 pKa = 10.78LEE1303 pKa = 4.16NVVTTDD1309 pKa = 3.34GWCVPAEE1316 pKa = 4.39GQDD1319 pKa = 3.57EE1320 pKa = 4.54ACRR1323 pKa = 11.84TVHH1326 pKa = 6.75INGAYY1331 pKa = 8.38TYY1333 pKa = 9.31SKK1335 pKa = 10.66SSEE1338 pKa = 4.31SPSGSLVRR1346 pKa = 11.84PGEE1349 pKa = 4.27VVTYY1353 pKa = 8.17TLEE1356 pKa = 4.12VEE1358 pKa = 4.77HH1359 pKa = 6.94VGTAPVSGAIVTDD1372 pKa = 4.36DD1373 pKa = 4.02LSQVLPYY1380 pKa = 10.94ASWNDD1385 pKa = 3.47DD1386 pKa = 3.39AEE1388 pKa = 4.53ATDD1391 pKa = 3.81GTVVRR1396 pKa = 11.84DD1397 pKa = 3.9GEE1399 pKa = 4.54TLTWTGDD1406 pKa = 3.66LAVGQVVEE1414 pKa = 4.11ITYY1417 pKa = 10.68SVTVGDD1423 pKa = 4.23LEE1425 pKa = 4.59EE1426 pKa = 4.46IAMVNVVTSPDD1437 pKa = 3.21EE1438 pKa = 4.03RR1439 pKa = 11.84AVCVPAADD1447 pKa = 4.25EE1448 pKa = 4.46NADD1451 pKa = 3.8CTTEE1455 pKa = 4.21HH1456 pKa = 6.72YY1457 pKa = 10.78NGAYY1461 pKa = 8.79TFSKK1465 pKa = 9.9TSDD1468 pKa = 3.62PASGSVVEE1476 pKa = 4.47AGEE1479 pKa = 4.34VVTYY1483 pKa = 8.45TVVVEE1488 pKa = 4.53HH1489 pKa = 6.74YY1490 pKa = 8.08GTAGVQDD1497 pKa = 5.23AIVVDD1502 pKa = 4.45DD1503 pKa = 4.0LTGVLAYY1510 pKa = 10.43AEE1512 pKa = 4.4WNDD1515 pKa = 4.28DD1516 pKa = 3.57AQADD1520 pKa = 4.11SGDD1523 pKa = 3.53LAYY1526 pKa = 10.37ADD1528 pKa = 5.01EE1529 pKa = 4.34EE1530 pKa = 4.7LVWSGDD1536 pKa = 3.54LAIGQVVEE1544 pKa = 3.84ITYY1547 pKa = 10.64SVTVGDD1553 pKa = 3.97EE1554 pKa = 4.3PEE1556 pKa = 4.12ATLHH1560 pKa = 6.12NVVTSPDD1567 pKa = 3.45DD1568 pKa = 3.46RR1569 pKa = 11.84AVCVPAADD1577 pKa = 4.28GNAGCATEE1585 pKa = 4.64HH1586 pKa = 5.53TTPPDD1591 pKa = 3.56APGKK1595 pKa = 9.45PPVPGLPSTGALLPWGMGLVALLLIACGGVLTITEE1630 pKa = 3.93QRR1632 pKa = 11.84KK1633 pKa = 7.82AASRR1637 pKa = 11.84ARR1639 pKa = 11.84QQIGTDD1645 pKa = 3.12RR1646 pKa = 11.84FF1647 pKa = 3.63
MM1 pKa = 7.44AVLIGVLGSMAMAVPVAADD20 pKa = 3.49TALEE24 pKa = 3.96PAIEE28 pKa = 4.75ADD30 pKa = 4.48SADD33 pKa = 3.74ADD35 pKa = 3.8ADD37 pKa = 3.74ADD39 pKa = 3.97ADD41 pKa = 3.74ADD43 pKa = 3.97AAADD47 pKa = 3.61AAAEE51 pKa = 4.12ADD53 pKa = 3.83STEE56 pKa = 4.02AAAEE60 pKa = 4.05EE61 pKa = 4.21DD62 pKa = 4.37TEE64 pKa = 4.5TLSDD68 pKa = 3.62TADD71 pKa = 3.36EE72 pKa = 4.54GTPATSSDD80 pKa = 3.91DD81 pKa = 3.58VSTLAEE87 pKa = 3.87VSPFAEE93 pKa = 4.35PSPVYY98 pKa = 10.21EE99 pKa = 4.01ITGVWLEE106 pKa = 4.04QPEE109 pKa = 4.55VTSRR113 pKa = 11.84GNPVVADD120 pKa = 3.15WRR122 pKa = 11.84INVNDD127 pKa = 3.99GNNPPSNEE135 pKa = 3.78LVDD138 pKa = 4.2NVVATFDD145 pKa = 3.19VGNAVFQTIPDD156 pKa = 3.79LCLTTGVQPASSISADD172 pKa = 3.59GSLLTCNFGTVALGRR187 pKa = 11.84SIALQTPVITEE198 pKa = 4.28GVTGTNVSLSGTSPAGEE215 pKa = 4.11TVEE218 pKa = 5.3LPLIPIQNDD227 pKa = 2.77FRR229 pKa = 11.84MDD231 pKa = 3.14MHH233 pKa = 7.16FGQNTNQRR241 pKa = 11.84EE242 pKa = 4.16WDD244 pKa = 3.79SLSNPTNVNLDD255 pKa = 3.85VQWSLRR261 pKa = 11.84LGKK264 pKa = 10.77GSDD267 pKa = 3.72PGPANQTFRR276 pKa = 11.84LRR278 pKa = 11.84IAPIFPAGSTVEE290 pKa = 4.34VGTHH294 pKa = 6.4IYY296 pKa = 9.35TGVVGCSPFDD306 pKa = 3.75LSNAPGHH313 pKa = 6.81PYY315 pKa = 10.46SSNPPHH321 pKa = 6.65PRR323 pKa = 11.84HH324 pKa = 5.89TNFVDD329 pKa = 3.41SCTLTAVPGEE339 pKa = 4.12PGVFDD344 pKa = 4.03LTLEE348 pKa = 4.64GINYY352 pKa = 10.0SLVNAPTNDD361 pKa = 3.2AGTANSPLPADD372 pKa = 3.55WDD374 pKa = 4.28YY375 pKa = 11.82VASGSLWFQVNAGSNGSLALTMDD398 pKa = 4.43PVTYY402 pKa = 9.18TATTGQTSEE411 pKa = 5.0DD412 pKa = 3.89LGTNNQTTKK421 pKa = 11.17AFTLPGAWSASFRR434 pKa = 11.84RR435 pKa = 11.84SATGSGGTFWDD446 pKa = 3.49DD447 pKa = 3.37SYY449 pKa = 11.78RR450 pKa = 11.84VTPGVTLRR458 pKa = 11.84NEE460 pKa = 3.75ARR462 pKa = 11.84NGYY465 pKa = 10.06SGDD468 pKa = 3.59RR469 pKa = 11.84NVAPTAQYY477 pKa = 10.82GSCLALDD484 pKa = 3.66TRR486 pKa = 11.84YY487 pKa = 10.88VIVDD491 pKa = 3.56RR492 pKa = 11.84DD493 pKa = 3.23FDD495 pKa = 4.23LAVATGFLARR505 pKa = 11.84NDD507 pKa = 3.62WPASAVLEE515 pKa = 4.44YY516 pKa = 10.92YY517 pKa = 10.88VGGSTLLDD525 pKa = 3.87PATGNTAYY533 pKa = 10.87NPDD536 pKa = 3.55QFEE539 pKa = 4.54CDD541 pKa = 3.66SDD543 pKa = 3.84PGGWTTEE550 pKa = 4.05LPADD554 pKa = 4.38LSTIRR559 pKa = 11.84AARR562 pKa = 11.84VTYY565 pKa = 10.27DD566 pKa = 3.48FSSFDD571 pKa = 3.33EE572 pKa = 4.35VGRR575 pKa = 11.84EE576 pKa = 4.09FIDD579 pKa = 3.65LVGWTTVRR587 pKa = 11.84EE588 pKa = 4.05DD589 pKa = 3.49VQPGQDD595 pKa = 2.07IWTFGSRR602 pKa = 11.84LRR604 pKa = 11.84NGGWFGPAQEE614 pKa = 4.57EE615 pKa = 5.39VITPTPDD622 pKa = 2.52ARR624 pKa = 11.84YY625 pKa = 8.07PHH627 pKa = 6.65TDD629 pKa = 2.44GRR631 pKa = 11.84RR632 pKa = 11.84DD633 pKa = 3.2IARR636 pKa = 11.84VISANPAIQKK646 pKa = 9.26SAEE649 pKa = 3.98RR650 pKa = 11.84SVVVPGVPTVFTLRR664 pKa = 11.84YY665 pKa = 8.55SANTGAGIPEE675 pKa = 4.35SVDD678 pKa = 2.96GYY680 pKa = 9.1TVVDD684 pKa = 3.81TLPPLVTYY692 pKa = 9.71EE693 pKa = 4.28VGSSDD698 pKa = 4.18PEE700 pKa = 4.05PTVTTNANNQQVLTWTFDD718 pKa = 3.57NVPTNVQNVLTYY730 pKa = 10.73AAAADD735 pKa = 3.9SSVAPGTPLVNTATASLGGTTSSPARR761 pKa = 11.84ATVTTANNGFTTILKK776 pKa = 9.07TSDD779 pKa = 3.1VEE781 pKa = 4.59FIPNRR786 pKa = 11.84LGDD789 pKa = 4.08GVGSGSWTVTIEE801 pKa = 4.03SRR803 pKa = 11.84DD804 pKa = 3.85PQSQAWTDD812 pKa = 3.68TIDD815 pKa = 3.21ILPYY819 pKa = 10.53NGDD822 pKa = 3.13GRR824 pKa = 11.84GTSFSGTYY832 pKa = 9.37TLGDD836 pKa = 3.27VVLFDD841 pKa = 5.17GGTLYY846 pKa = 9.82YY847 pKa = 10.43TDD849 pKa = 5.36ADD851 pKa = 4.23PATLSDD857 pKa = 4.56DD858 pKa = 4.08PAHH861 pKa = 6.79EE862 pKa = 4.96SNGAPNAPSALWSTEE877 pKa = 3.66RR878 pKa = 11.84PEE880 pKa = 4.12NPTAIRR886 pKa = 11.84VIGGTLATGGSFSFQVPITTDD907 pKa = 3.02GAEE910 pKa = 4.15AQDD913 pKa = 3.61VFVNRR918 pKa = 11.84AQARR922 pKa = 11.84AEE924 pKa = 3.91HH925 pKa = 6.5TEE927 pKa = 3.72LVMRR931 pKa = 11.84TSAALVVTDD940 pKa = 3.55YY941 pKa = 11.32LVEE944 pKa = 4.24KK945 pKa = 10.22SANPEE950 pKa = 3.7SGSTVRR956 pKa = 11.84PGDD959 pKa = 3.58VVEE962 pKa = 4.26YY963 pKa = 9.29TVTVSQMGDD972 pKa = 3.17VPAGAWFSDD981 pKa = 3.68DD982 pKa = 3.71MSDD985 pKa = 4.18VLDD988 pKa = 4.42DD989 pKa = 4.02ADD991 pKa = 3.99YY992 pKa = 11.51NGDD995 pKa = 3.17VTASHH1000 pKa = 6.79GEE1002 pKa = 3.97VLSFEE1007 pKa = 4.28NGILEE1012 pKa = 4.37WEE1014 pKa = 4.6GVVPVGEE1021 pKa = 4.35IATITYY1027 pKa = 10.29SVTVKK1032 pKa = 10.44AAEE1035 pKa = 4.09EE1036 pKa = 3.85LRR1038 pKa = 11.84EE1039 pKa = 4.13GDD1041 pKa = 4.7SNTFLNNQVTSPGCQGACSTEE1062 pKa = 4.02HH1063 pKa = 6.03LVGYY1067 pKa = 9.57YY1068 pKa = 9.96SVSKK1072 pKa = 9.09TAEE1075 pKa = 4.03PAPGEE1080 pKa = 4.19TVRR1083 pKa = 11.84IGDD1086 pKa = 3.87VITYY1090 pKa = 8.78HH1091 pKa = 5.79VQVTQHH1097 pKa = 6.42GEE1099 pKa = 4.1GAVDD1103 pKa = 3.2NAFFIDD1109 pKa = 4.52DD1110 pKa = 4.15LSEE1113 pKa = 4.15VLDD1116 pKa = 3.96DD1117 pKa = 6.57AIWNDD1122 pKa = 3.64DD1123 pKa = 3.32AAATDD1128 pKa = 4.39GEE1130 pKa = 4.57LSFDD1134 pKa = 3.88EE1135 pKa = 6.13PILSWSAPLEE1145 pKa = 4.32VGDD1148 pKa = 4.39VVDD1151 pKa = 3.37VTYY1154 pKa = 11.08SVTVTGEE1161 pKa = 3.49GDD1163 pKa = 3.5YY1164 pKa = 11.05EE1165 pKa = 4.1LRR1167 pKa = 11.84NVVSSVCAEE1176 pKa = 4.62DD1177 pKa = 6.06DD1178 pKa = 4.05PDD1180 pKa = 4.21CLEE1183 pKa = 5.05AICVPAADD1191 pKa = 4.8EE1192 pKa = 5.12NPDD1195 pKa = 3.53CTTEE1199 pKa = 4.58HH1200 pKa = 6.59IVGDD1204 pKa = 3.86YY1205 pKa = 10.63QVTKK1209 pKa = 10.9VSDD1212 pKa = 4.07PEE1214 pKa = 4.14SGSAVEE1220 pKa = 4.34VGDD1223 pKa = 4.0VIDD1226 pKa = 3.76YY1227 pKa = 7.79TVTVSHH1233 pKa = 7.04IGLADD1238 pKa = 3.56VEE1240 pKa = 4.41ASFVDD1245 pKa = 3.97DD1246 pKa = 5.32LSDD1249 pKa = 4.22VLDD1252 pKa = 4.09DD1253 pKa = 6.15AIWNDD1258 pKa = 4.05DD1259 pKa = 3.28VSASAGEE1266 pKa = 4.04FSFEE1270 pKa = 4.67GEE1272 pKa = 4.21TLSWWGALSEE1282 pKa = 4.31GDD1284 pKa = 4.0VVTVTYY1290 pKa = 10.65SVTVTAEE1297 pKa = 3.63GDD1299 pKa = 3.38RR1300 pKa = 11.84YY1301 pKa = 10.78LEE1303 pKa = 4.16NVVTTDD1309 pKa = 3.34GWCVPAEE1316 pKa = 4.39GQDD1319 pKa = 3.57EE1320 pKa = 4.54ACRR1323 pKa = 11.84TVHH1326 pKa = 6.75INGAYY1331 pKa = 8.38TYY1333 pKa = 9.31SKK1335 pKa = 10.66SSEE1338 pKa = 4.31SPSGSLVRR1346 pKa = 11.84PGEE1349 pKa = 4.27VVTYY1353 pKa = 8.17TLEE1356 pKa = 4.12VEE1358 pKa = 4.77HH1359 pKa = 6.94VGTAPVSGAIVTDD1372 pKa = 4.36DD1373 pKa = 4.02LSQVLPYY1380 pKa = 10.94ASWNDD1385 pKa = 3.47DD1386 pKa = 3.39AEE1388 pKa = 4.53ATDD1391 pKa = 3.81GTVVRR1396 pKa = 11.84DD1397 pKa = 3.9GEE1399 pKa = 4.54TLTWTGDD1406 pKa = 3.66LAVGQVVEE1414 pKa = 4.11ITYY1417 pKa = 10.68SVTVGDD1423 pKa = 4.23LEE1425 pKa = 4.59EE1426 pKa = 4.46IAMVNVVTSPDD1437 pKa = 3.21EE1438 pKa = 4.03RR1439 pKa = 11.84AVCVPAADD1447 pKa = 4.25EE1448 pKa = 4.46NADD1451 pKa = 3.8CTTEE1455 pKa = 4.21HH1456 pKa = 6.72YY1457 pKa = 10.78NGAYY1461 pKa = 8.79TFSKK1465 pKa = 9.9TSDD1468 pKa = 3.62PASGSVVEE1476 pKa = 4.47AGEE1479 pKa = 4.34VVTYY1483 pKa = 8.45TVVVEE1488 pKa = 4.53HH1489 pKa = 6.74YY1490 pKa = 8.08GTAGVQDD1497 pKa = 5.23AIVVDD1502 pKa = 4.45DD1503 pKa = 4.0LTGVLAYY1510 pKa = 10.43AEE1512 pKa = 4.4WNDD1515 pKa = 4.28DD1516 pKa = 3.57AQADD1520 pKa = 4.11SGDD1523 pKa = 3.53LAYY1526 pKa = 10.37ADD1528 pKa = 5.01EE1529 pKa = 4.34EE1530 pKa = 4.7LVWSGDD1536 pKa = 3.54LAIGQVVEE1544 pKa = 3.84ITYY1547 pKa = 10.64SVTVGDD1553 pKa = 3.97EE1554 pKa = 4.3PEE1556 pKa = 4.12ATLHH1560 pKa = 6.12NVVTSPDD1567 pKa = 3.45DD1568 pKa = 3.46RR1569 pKa = 11.84AVCVPAADD1577 pKa = 4.28GNAGCATEE1585 pKa = 4.64HH1586 pKa = 5.53TTPPDD1591 pKa = 3.56APGKK1595 pKa = 9.45PPVPGLPSTGALLPWGMGLVALLLIACGGVLTITEE1630 pKa = 3.93QRR1632 pKa = 11.84KK1633 pKa = 7.82AASRR1637 pKa = 11.84ARR1639 pKa = 11.84QQIGTDD1645 pKa = 3.12RR1646 pKa = 11.84FF1647 pKa = 3.63
Molecular weight: 173.21 kDa
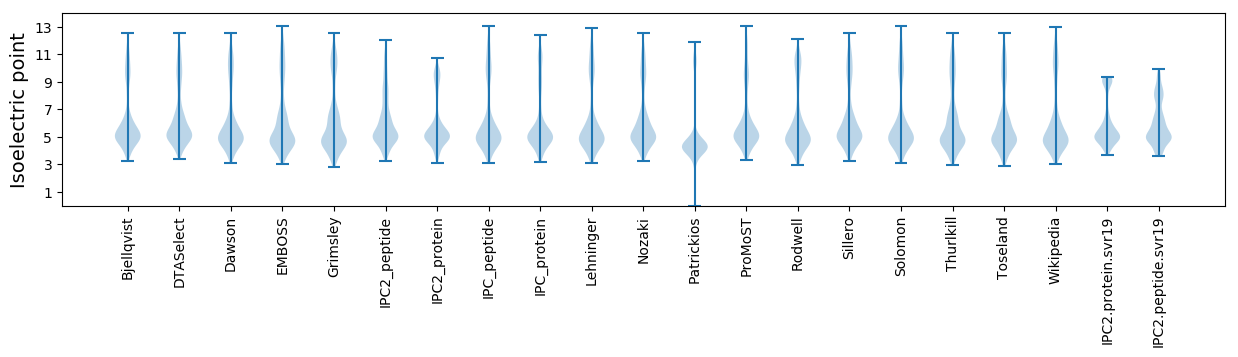
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A3N2BG53|A0A3N2BG53_9MICO 23S rRNA (Cytidine1920-2'-O)/16S rRNA (Cytidine1409-2'-O)-methyltransferase OS=Bogoriella caseilytica OX=56055 GN=EDD31_2631 PE=4 SV=1
MM1 pKa = 7.3ARR3 pKa = 11.84AARR6 pKa = 11.84GNIVLALLALLALLALLALLALLALLALLALARR39 pKa = 11.84PAGTSRR45 pKa = 11.84LGAATSGPTRR55 pKa = 11.84RR56 pKa = 11.84ATPHH60 pKa = 6.69RR61 pKa = 11.84LPRR64 pKa = 11.84DD65 pKa = 3.37AGGVRR70 pKa = 3.61
MM1 pKa = 7.3ARR3 pKa = 11.84AARR6 pKa = 11.84GNIVLALLALLALLALLALLALLALLALLALARR39 pKa = 11.84PAGTSRR45 pKa = 11.84LGAATSGPTRR55 pKa = 11.84RR56 pKa = 11.84ATPHH60 pKa = 6.69RR61 pKa = 11.84LPRR64 pKa = 11.84DD65 pKa = 3.37AGGVRR70 pKa = 3.61
Molecular weight: 7.08 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
992085 |
30 |
2576 |
348.8 |
37.33 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
13.521 ± 0.061 | 0.539 ± 0.011 |
5.789 ± 0.043 | 6.728 ± 0.051 |
2.756 ± 0.03 | 9.151 ± 0.041 |
2.257 ± 0.024 | 3.916 ± 0.028 |
1.493 ± 0.03 | 10.372 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.922 ± 0.019 | 1.78 ± 0.022 |
5.721 ± 0.04 | 3.088 ± 0.024 |
7.372 ± 0.058 | 5.59 ± 0.029 |
5.897 ± 0.038 | 8.62 ± 0.043 |
1.539 ± 0.019 | 1.949 ± 0.022 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
