
Human papillomavirus type 5b
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cossaviricota; Papovaviricetes; Zurhausenvirales; Papillomaviridae; Firstpapillomavirinae; Betapapillomavirus; Betapapillomavirus 1
Average proteome isoelectric point is 6.55
Get precalculated fractions of proteins
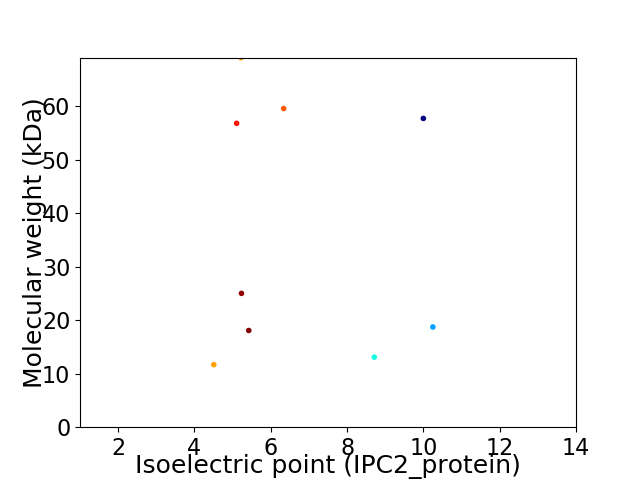
Virtual 2D-PAGE plot for 9 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>sp|P26559|VE7_HPV5B Protein E7 OS=Human papillomavirus type 5b OX=10599 GN=E7 PE=3 SV=1
MM1 pKa = 7.41IGKK4 pKa = 8.9EE5 pKa = 4.0VTVQDD10 pKa = 4.38IILEE14 pKa = 4.05LSEE17 pKa = 4.23VQPEE21 pKa = 4.28VLPVDD26 pKa = 4.17LFCEE30 pKa = 4.15EE31 pKa = 4.45EE32 pKa = 4.38LPNEE36 pKa = 4.16QEE38 pKa = 4.26TEE40 pKa = 4.09EE41 pKa = 4.39EE42 pKa = 3.92PDD44 pKa = 3.26IEE46 pKa = 4.74RR47 pKa = 11.84ISYY50 pKa = 9.99KK51 pKa = 10.66VIAPCGCRR59 pKa = 11.84HH60 pKa = 6.38CEE62 pKa = 3.5VKK64 pKa = 10.77LRR66 pKa = 11.84IFVHH70 pKa = 5.08ATEE73 pKa = 4.28FGIRR77 pKa = 11.84AFQQLLTGDD86 pKa = 4.25LQLLCPDD93 pKa = 3.44CRR95 pKa = 11.84GNCKK99 pKa = 9.94HH100 pKa = 7.02DD101 pKa = 3.47GSS103 pKa = 4.21
MM1 pKa = 7.41IGKK4 pKa = 8.9EE5 pKa = 4.0VTVQDD10 pKa = 4.38IILEE14 pKa = 4.05LSEE17 pKa = 4.23VQPEE21 pKa = 4.28VLPVDD26 pKa = 4.17LFCEE30 pKa = 4.15EE31 pKa = 4.45EE32 pKa = 4.38LPNEE36 pKa = 4.16QEE38 pKa = 4.26TEE40 pKa = 4.09EE41 pKa = 4.39EE42 pKa = 3.92PDD44 pKa = 3.26IEE46 pKa = 4.74RR47 pKa = 11.84ISYY50 pKa = 9.99KK51 pKa = 10.66VIAPCGCRR59 pKa = 11.84HH60 pKa = 6.38CEE62 pKa = 3.5VKK64 pKa = 10.77LRR66 pKa = 11.84IFVHH70 pKa = 5.08ATEE73 pKa = 4.28FGIRR77 pKa = 11.84AFQQLLTGDD86 pKa = 4.25LQLLCPDD93 pKa = 3.44CRR95 pKa = 11.84GNCKK99 pKa = 9.94HH100 pKa = 7.02DD101 pKa = 3.47GSS103 pKa = 4.21
Molecular weight: 11.7 kDa
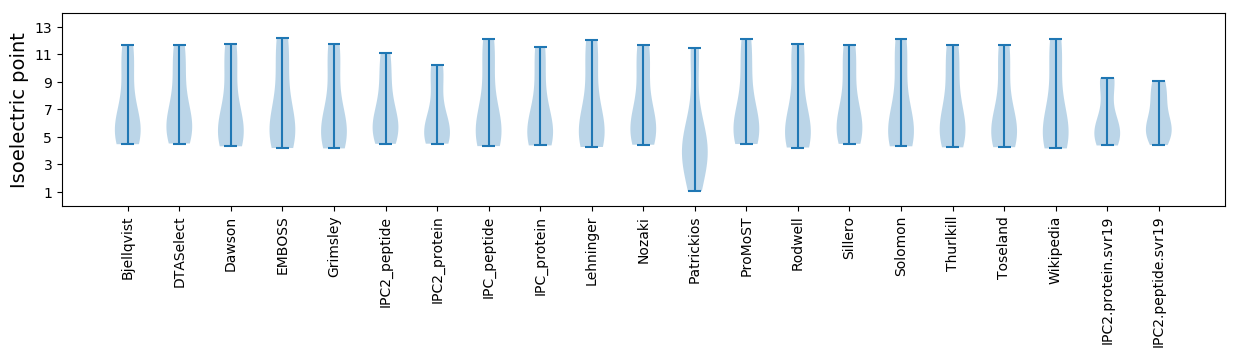
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|P26550|VE4_HPV5B Protein E4 OS=Human papillomavirus type 5b OX=10599 GN=E4 PE=3 SV=2
MM1 pKa = 7.68EE2 pKa = 5.02NLSEE6 pKa = 4.76RR7 pKa = 11.84FNALQDD13 pKa = 3.48QLMNIYY19 pKa = 9.48EE20 pKa = 4.27AAEE23 pKa = 3.98QTLQAQIKK31 pKa = 8.71HH32 pKa = 4.61WQTLRR37 pKa = 11.84KK38 pKa = 9.06EE39 pKa = 4.28AVLLYY44 pKa = 8.61YY45 pKa = 10.61ARR47 pKa = 11.84EE48 pKa = 4.03KK49 pKa = 11.0GVTRR53 pKa = 11.84LGYY56 pKa = 10.26QPVPVKK62 pKa = 10.56AVSEE66 pKa = 4.56TKK68 pKa = 10.5AKK70 pKa = 10.15EE71 pKa = 4.07AIAMVLQLEE80 pKa = 4.42SLQTSDD86 pKa = 4.56FAHH89 pKa = 6.46EE90 pKa = 4.1PWTLVDD96 pKa = 3.67TSTEE100 pKa = 4.25TFRR103 pKa = 11.84SAPEE107 pKa = 3.64GHH109 pKa = 6.37FKK111 pKa = 10.66KK112 pKa = 10.8GPVPVEE118 pKa = 3.74VIYY121 pKa = 11.25DD122 pKa = 3.85NDD124 pKa = 3.86PDD126 pKa = 3.89NANLYY131 pKa = 8.02TMWTYY136 pKa = 11.13VYY138 pKa = 11.46YY139 pKa = 10.27MDD141 pKa = 6.38ADD143 pKa = 4.42DD144 pKa = 4.48KK145 pKa = 9.72WHH147 pKa = 6.99KK148 pKa = 10.57ARR150 pKa = 11.84SGVNHH155 pKa = 6.5IGIYY159 pKa = 9.54YY160 pKa = 9.95LQGTFKK166 pKa = 10.86NYY168 pKa = 10.12YY169 pKa = 10.21VLFADD174 pKa = 3.83DD175 pKa = 3.8AKK177 pKa = 10.94RR178 pKa = 11.84YY179 pKa = 6.43GTTGEE184 pKa = 4.16WEE186 pKa = 4.22VKK188 pKa = 9.71VNKK191 pKa = 9.12DD192 pKa = 3.24TVFAPVTSSTPPGSPGRR209 pKa = 11.84QADD212 pKa = 3.55TDD214 pKa = 4.05TTAKK218 pKa = 10.05TPTTSTTAVDD228 pKa = 3.52STSRR232 pKa = 11.84QLTTSKK238 pKa = 10.35QPQQTEE244 pKa = 4.12TRR246 pKa = 11.84GRR248 pKa = 11.84RR249 pKa = 11.84YY250 pKa = 10.09GRR252 pKa = 11.84RR253 pKa = 11.84PSSKK257 pKa = 9.79SRR259 pKa = 11.84RR260 pKa = 11.84SQTQQRR266 pKa = 11.84RR267 pKa = 11.84SRR269 pKa = 11.84SRR271 pKa = 11.84HH272 pKa = 4.65RR273 pKa = 11.84SRR275 pKa = 11.84SRR277 pKa = 11.84SRR279 pKa = 11.84SRR281 pKa = 11.84SKK283 pKa = 10.9SQTHH287 pKa = 5.19TTWSTTRR294 pKa = 11.84SRR296 pKa = 11.84STSVGKK302 pKa = 8.3TRR304 pKa = 11.84ALTSRR309 pKa = 11.84SRR311 pKa = 11.84SRR313 pKa = 11.84GRR315 pKa = 11.84SPSTCRR321 pKa = 11.84RR322 pKa = 11.84GGGRR326 pKa = 11.84SPRR329 pKa = 11.84RR330 pKa = 11.84RR331 pKa = 11.84SRR333 pKa = 11.84SPSTYY338 pKa = 10.42SSCTTQRR345 pKa = 11.84SQRR348 pKa = 11.84ARR350 pKa = 11.84AEE352 pKa = 4.19SPTTRR357 pKa = 11.84GARR360 pKa = 11.84GSRR363 pKa = 11.84GSRR366 pKa = 11.84GGSRR370 pKa = 11.84GGRR373 pKa = 11.84LRR375 pKa = 11.84RR376 pKa = 11.84RR377 pKa = 11.84GRR379 pKa = 11.84SSSSSSPAHH388 pKa = 5.78KK389 pKa = 10.08RR390 pKa = 11.84SRR392 pKa = 11.84GGSAKK397 pKa = 10.31LRR399 pKa = 11.84GVSPGEE405 pKa = 3.81VGGSLRR411 pKa = 11.84SVSSKK416 pKa = 9.03HH417 pKa = 4.63TGRR420 pKa = 11.84LGRR423 pKa = 11.84LLEE426 pKa = 4.17EE427 pKa = 4.57ARR429 pKa = 11.84DD430 pKa = 3.72PPVIIVKK437 pKa = 10.23GAANTLKK444 pKa = 10.56YY445 pKa = 10.26FRR447 pKa = 11.84NRR449 pKa = 11.84AKK451 pKa = 10.21IKK453 pKa = 8.54YY454 pKa = 7.56TGLFRR459 pKa = 11.84SFSTTWSWVAGDD471 pKa = 3.34GTEE474 pKa = 3.79RR475 pKa = 11.84LGRR478 pKa = 11.84PRR480 pKa = 11.84MLISFSSYY488 pKa = 7.26SQRR491 pKa = 11.84RR492 pKa = 11.84DD493 pKa = 2.62FDD495 pKa = 3.58EE496 pKa = 4.1AVRR499 pKa = 11.84YY500 pKa = 8.12PKK502 pKa = 10.76GVDD505 pKa = 2.9KK506 pKa = 11.05AYY508 pKa = 10.91GNLDD512 pKa = 3.48SLL514 pKa = 4.64
MM1 pKa = 7.68EE2 pKa = 5.02NLSEE6 pKa = 4.76RR7 pKa = 11.84FNALQDD13 pKa = 3.48QLMNIYY19 pKa = 9.48EE20 pKa = 4.27AAEE23 pKa = 3.98QTLQAQIKK31 pKa = 8.71HH32 pKa = 4.61WQTLRR37 pKa = 11.84KK38 pKa = 9.06EE39 pKa = 4.28AVLLYY44 pKa = 8.61YY45 pKa = 10.61ARR47 pKa = 11.84EE48 pKa = 4.03KK49 pKa = 11.0GVTRR53 pKa = 11.84LGYY56 pKa = 10.26QPVPVKK62 pKa = 10.56AVSEE66 pKa = 4.56TKK68 pKa = 10.5AKK70 pKa = 10.15EE71 pKa = 4.07AIAMVLQLEE80 pKa = 4.42SLQTSDD86 pKa = 4.56FAHH89 pKa = 6.46EE90 pKa = 4.1PWTLVDD96 pKa = 3.67TSTEE100 pKa = 4.25TFRR103 pKa = 11.84SAPEE107 pKa = 3.64GHH109 pKa = 6.37FKK111 pKa = 10.66KK112 pKa = 10.8GPVPVEE118 pKa = 3.74VIYY121 pKa = 11.25DD122 pKa = 3.85NDD124 pKa = 3.86PDD126 pKa = 3.89NANLYY131 pKa = 8.02TMWTYY136 pKa = 11.13VYY138 pKa = 11.46YY139 pKa = 10.27MDD141 pKa = 6.38ADD143 pKa = 4.42DD144 pKa = 4.48KK145 pKa = 9.72WHH147 pKa = 6.99KK148 pKa = 10.57ARR150 pKa = 11.84SGVNHH155 pKa = 6.5IGIYY159 pKa = 9.54YY160 pKa = 9.95LQGTFKK166 pKa = 10.86NYY168 pKa = 10.12YY169 pKa = 10.21VLFADD174 pKa = 3.83DD175 pKa = 3.8AKK177 pKa = 10.94RR178 pKa = 11.84YY179 pKa = 6.43GTTGEE184 pKa = 4.16WEE186 pKa = 4.22VKK188 pKa = 9.71VNKK191 pKa = 9.12DD192 pKa = 3.24TVFAPVTSSTPPGSPGRR209 pKa = 11.84QADD212 pKa = 3.55TDD214 pKa = 4.05TTAKK218 pKa = 10.05TPTTSTTAVDD228 pKa = 3.52STSRR232 pKa = 11.84QLTTSKK238 pKa = 10.35QPQQTEE244 pKa = 4.12TRR246 pKa = 11.84GRR248 pKa = 11.84RR249 pKa = 11.84YY250 pKa = 10.09GRR252 pKa = 11.84RR253 pKa = 11.84PSSKK257 pKa = 9.79SRR259 pKa = 11.84RR260 pKa = 11.84SQTQQRR266 pKa = 11.84RR267 pKa = 11.84SRR269 pKa = 11.84SRR271 pKa = 11.84HH272 pKa = 4.65RR273 pKa = 11.84SRR275 pKa = 11.84SRR277 pKa = 11.84SRR279 pKa = 11.84SRR281 pKa = 11.84SKK283 pKa = 10.9SQTHH287 pKa = 5.19TTWSTTRR294 pKa = 11.84SRR296 pKa = 11.84STSVGKK302 pKa = 8.3TRR304 pKa = 11.84ALTSRR309 pKa = 11.84SRR311 pKa = 11.84SRR313 pKa = 11.84GRR315 pKa = 11.84SPSTCRR321 pKa = 11.84RR322 pKa = 11.84GGGRR326 pKa = 11.84SPRR329 pKa = 11.84RR330 pKa = 11.84RR331 pKa = 11.84SRR333 pKa = 11.84SPSTYY338 pKa = 10.42SSCTTQRR345 pKa = 11.84SQRR348 pKa = 11.84ARR350 pKa = 11.84AEE352 pKa = 4.19SPTTRR357 pKa = 11.84GARR360 pKa = 11.84GSRR363 pKa = 11.84GSRR366 pKa = 11.84GGSRR370 pKa = 11.84GGRR373 pKa = 11.84LRR375 pKa = 11.84RR376 pKa = 11.84RR377 pKa = 11.84GRR379 pKa = 11.84SSSSSSPAHH388 pKa = 5.78KK389 pKa = 10.08RR390 pKa = 11.84SRR392 pKa = 11.84GGSAKK397 pKa = 10.31LRR399 pKa = 11.84GVSPGEE405 pKa = 3.81VGGSLRR411 pKa = 11.84SVSSKK416 pKa = 9.03HH417 pKa = 4.63TGRR420 pKa = 11.84LGRR423 pKa = 11.84LLEE426 pKa = 4.17EE427 pKa = 4.57ARR429 pKa = 11.84DD430 pKa = 3.72PPVIIVKK437 pKa = 10.23GAANTLKK444 pKa = 10.56YY445 pKa = 10.26FRR447 pKa = 11.84NRR449 pKa = 11.84AKK451 pKa = 10.21IKK453 pKa = 8.54YY454 pKa = 7.56TGLFRR459 pKa = 11.84SFSTTWSWVAGDD471 pKa = 3.34GTEE474 pKa = 3.79RR475 pKa = 11.84LGRR478 pKa = 11.84PRR480 pKa = 11.84MLISFSSYY488 pKa = 7.26SQRR491 pKa = 11.84RR492 pKa = 11.84DD493 pKa = 2.62FDD495 pKa = 3.58EE496 pKa = 4.1AVRR499 pKa = 11.84YY500 pKa = 8.12PKK502 pKa = 10.76GVDD505 pKa = 2.9KK506 pKa = 11.05AYY508 pKa = 10.91GNLDD512 pKa = 3.48SLL514 pKa = 4.64
Molecular weight: 57.75 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2941 |
103 |
606 |
326.8 |
36.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.712 ± 0.432 | 1.904 ± 0.604 |
5.712 ± 0.615 | 5.916 ± 0.687 |
3.706 ± 0.605 | 7.31 ± 0.851 |
2.89 ± 0.489 | 4.556 ± 0.736 |
5.236 ± 0.684 | 7.718 ± 0.688 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.36 ± 0.361 | 3.876 ± 0.62 |
6.936 ± 1.551 | 5.168 ± 0.387 |
6.936 ± 1.325 | 7.446 ± 1.212 |
6.834 ± 1.014 | 6.188 ± 0.9 |
1.292 ± 0.298 | 3.298 ± 0.445 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
