
Circovirus-like genome RW-D
Taxonomy: Viruses; unclassified viruses; unclassified DNA viruses; unclassified ssDNA viruses
Average proteome isoelectric point is 8.07
Get precalculated fractions of proteins
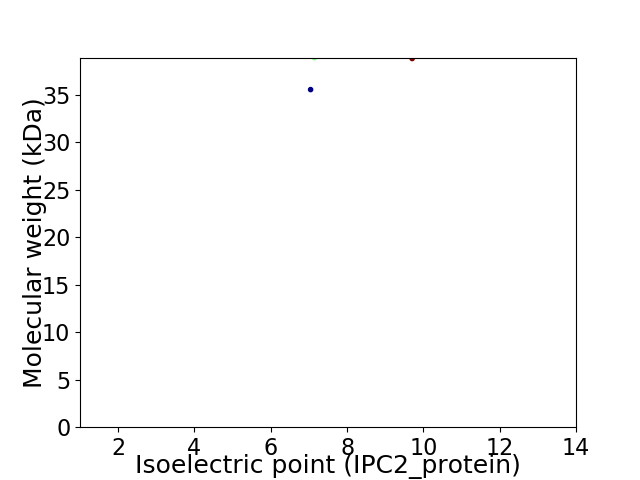
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|C6GII2|C6GII2_9VIRU ATP-dependent helicase Rep OS=Circovirus-like genome RW-D OX=642254 PE=3 SV=1
MM1 pKa = 7.56LAQAPGRR8 pKa = 11.84YY9 pKa = 8.38RR10 pKa = 11.84NWVITVNNWTEE21 pKa = 3.73NDD23 pKa = 3.41YY24 pKa = 11.5KK25 pKa = 10.99LALLSPYY32 pKa = 9.86RR33 pKa = 11.84YY34 pKa = 9.45IIIGRR39 pKa = 11.84EE40 pKa = 3.58RR41 pKa = 11.84GEE43 pKa = 4.45CNTPHH48 pKa = 5.99LQIYY52 pKa = 8.59LQLYY56 pKa = 7.88HH57 pKa = 7.12AKK59 pKa = 10.6SFDD62 pKa = 3.55TVKK65 pKa = 10.92QNFFPRR71 pKa = 11.84AHH73 pKa = 6.79LEE75 pKa = 4.06ASHH78 pKa = 6.33SKK80 pKa = 9.66PRR82 pKa = 11.84QARR85 pKa = 11.84AYY87 pKa = 7.14CTKK90 pKa = 10.01EE91 pKa = 3.54HH92 pKa = 5.85YY93 pKa = 10.25FEE95 pKa = 4.77YY96 pKa = 11.37GEE98 pKa = 4.65MSTQGKK104 pKa = 7.89RR105 pKa = 11.84TDD107 pKa = 3.47LQVAQEE113 pKa = 4.53LLDD116 pKa = 3.7SGISIRR122 pKa = 11.84QALEE126 pKa = 3.82SEE128 pKa = 5.22MITSMGALAAYY139 pKa = 7.87EE140 pKa = 4.29KK141 pKa = 10.11LQKK144 pKa = 10.51YY145 pKa = 6.5YY146 pKa = 9.75TVHH149 pKa = 6.6RR150 pKa = 11.84PRR152 pKa = 11.84PRR154 pKa = 11.84VVWIYY159 pKa = 11.22GPAGSGKK166 pKa = 7.72TDD168 pKa = 3.02KK169 pKa = 10.63AYY171 pKa = 10.73SISGPDD177 pKa = 3.36VYY179 pKa = 11.06KK180 pKa = 10.84SDD182 pKa = 4.78LIKK185 pKa = 10.85EE186 pKa = 4.36GWFDD190 pKa = 3.46GYY192 pKa = 11.11DD193 pKa = 3.08RR194 pKa = 11.84HH195 pKa = 7.13RR196 pKa = 11.84SIIIDD201 pKa = 3.78DD202 pKa = 4.28LEE204 pKa = 4.99IDD206 pKa = 4.06RR207 pKa = 11.84DD208 pKa = 3.97DD209 pKa = 4.06KK210 pKa = 11.31KK211 pKa = 9.54TFGLLLSLLDD221 pKa = 4.42KK222 pKa = 11.09NPQKK226 pKa = 11.4VNVKK230 pKa = 9.99GSSASILADD239 pKa = 3.61TIVITCQQAPWHH251 pKa = 6.01IWYY254 pKa = 9.3HH255 pKa = 5.99PSDD258 pKa = 3.89NMSLPFKK265 pKa = 10.91HH266 pKa = 5.7MATRR270 pKa = 11.84EE271 pKa = 3.98EE272 pKa = 4.06IEE274 pKa = 4.1RR275 pKa = 11.84DD276 pKa = 2.94VDD278 pKa = 3.28LRR280 pKa = 11.84QIMRR284 pKa = 11.84RR285 pKa = 11.84ITEE288 pKa = 4.69IIHH291 pKa = 6.23LTDD294 pKa = 3.29TDD296 pKa = 3.7KK297 pKa = 11.56VKK299 pKa = 10.77YY300 pKa = 10.23PEE302 pKa = 4.03VTEE305 pKa = 4.22VV306 pKa = 2.91
MM1 pKa = 7.56LAQAPGRR8 pKa = 11.84YY9 pKa = 8.38RR10 pKa = 11.84NWVITVNNWTEE21 pKa = 3.73NDD23 pKa = 3.41YY24 pKa = 11.5KK25 pKa = 10.99LALLSPYY32 pKa = 9.86RR33 pKa = 11.84YY34 pKa = 9.45IIIGRR39 pKa = 11.84EE40 pKa = 3.58RR41 pKa = 11.84GEE43 pKa = 4.45CNTPHH48 pKa = 5.99LQIYY52 pKa = 8.59LQLYY56 pKa = 7.88HH57 pKa = 7.12AKK59 pKa = 10.6SFDD62 pKa = 3.55TVKK65 pKa = 10.92QNFFPRR71 pKa = 11.84AHH73 pKa = 6.79LEE75 pKa = 4.06ASHH78 pKa = 6.33SKK80 pKa = 9.66PRR82 pKa = 11.84QARR85 pKa = 11.84AYY87 pKa = 7.14CTKK90 pKa = 10.01EE91 pKa = 3.54HH92 pKa = 5.85YY93 pKa = 10.25FEE95 pKa = 4.77YY96 pKa = 11.37GEE98 pKa = 4.65MSTQGKK104 pKa = 7.89RR105 pKa = 11.84TDD107 pKa = 3.47LQVAQEE113 pKa = 4.53LLDD116 pKa = 3.7SGISIRR122 pKa = 11.84QALEE126 pKa = 3.82SEE128 pKa = 5.22MITSMGALAAYY139 pKa = 7.87EE140 pKa = 4.29KK141 pKa = 10.11LQKK144 pKa = 10.51YY145 pKa = 6.5YY146 pKa = 9.75TVHH149 pKa = 6.6RR150 pKa = 11.84PRR152 pKa = 11.84PRR154 pKa = 11.84VVWIYY159 pKa = 11.22GPAGSGKK166 pKa = 7.72TDD168 pKa = 3.02KK169 pKa = 10.63AYY171 pKa = 10.73SISGPDD177 pKa = 3.36VYY179 pKa = 11.06KK180 pKa = 10.84SDD182 pKa = 4.78LIKK185 pKa = 10.85EE186 pKa = 4.36GWFDD190 pKa = 3.46GYY192 pKa = 11.11DD193 pKa = 3.08RR194 pKa = 11.84HH195 pKa = 7.13RR196 pKa = 11.84SIIIDD201 pKa = 3.78DD202 pKa = 4.28LEE204 pKa = 4.99IDD206 pKa = 4.06RR207 pKa = 11.84DD208 pKa = 3.97DD209 pKa = 4.06KK210 pKa = 11.31KK211 pKa = 9.54TFGLLLSLLDD221 pKa = 4.42KK222 pKa = 11.09NPQKK226 pKa = 11.4VNVKK230 pKa = 9.99GSSASILADD239 pKa = 3.61TIVITCQQAPWHH251 pKa = 6.01IWYY254 pKa = 9.3HH255 pKa = 5.99PSDD258 pKa = 3.89NMSLPFKK265 pKa = 10.91HH266 pKa = 5.7MATRR270 pKa = 11.84EE271 pKa = 3.98EE272 pKa = 4.06IEE274 pKa = 4.1RR275 pKa = 11.84DD276 pKa = 2.94VDD278 pKa = 3.28LRR280 pKa = 11.84QIMRR284 pKa = 11.84RR285 pKa = 11.84ITEE288 pKa = 4.69IIHH291 pKa = 6.23LTDD294 pKa = 3.29TDD296 pKa = 3.7KK297 pKa = 11.56VKK299 pKa = 10.77YY300 pKa = 10.23PEE302 pKa = 4.03VTEE305 pKa = 4.22VV306 pKa = 2.91
Molecular weight: 35.53 kDa
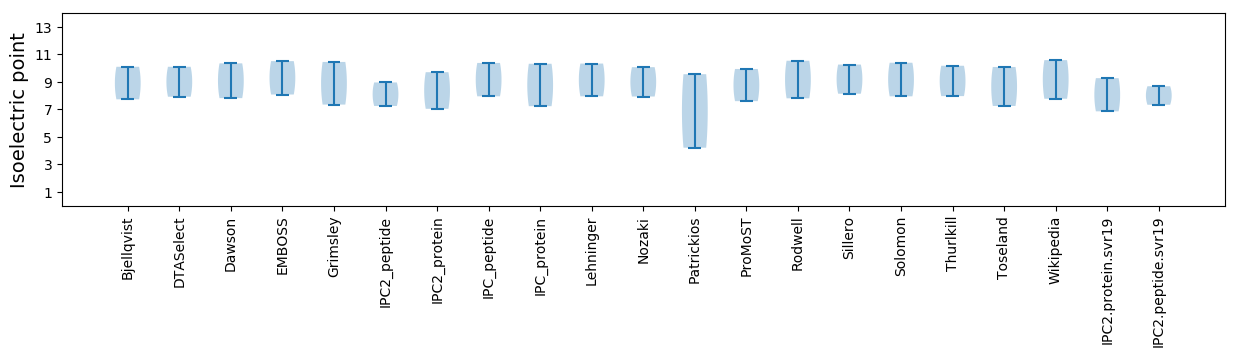
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|C6GII2|C6GII2_9VIRU ATP-dependent helicase Rep OS=Circovirus-like genome RW-D OX=642254 PE=3 SV=1
MM1 pKa = 7.79RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84PAYY7 pKa = 9.96VGPSWGGSRR16 pKa = 11.84IGGANIPMNTRR27 pKa = 11.84AKK29 pKa = 9.85RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 9.6IIPLLWNSGSKK45 pKa = 9.9LANNLNLGFGSRR57 pKa = 11.84GRR59 pKa = 11.84GPGSMVSRR67 pKa = 11.84IRR69 pKa = 11.84NYY71 pKa = 10.82KK72 pKa = 9.19MEE74 pKa = 4.48ISVKK78 pKa = 9.99PVGTGSSYY86 pKa = 11.02SYY88 pKa = 10.68YY89 pKa = 10.45RR90 pKa = 11.84YY91 pKa = 8.07TRR93 pKa = 11.84RR94 pKa = 11.84PDD96 pKa = 2.87KK97 pKa = 10.71GSRR100 pKa = 11.84IVKK103 pKa = 7.41TQQAPLFSVFNSGSRR118 pKa = 11.84MTSLQGEE125 pKa = 4.25QGYY128 pKa = 6.47TTLSVMTGAKK138 pKa = 9.57LRR140 pKa = 11.84SLYY143 pKa = 11.04LEE145 pKa = 4.34TSANQDD151 pKa = 2.46GRR153 pKa = 11.84FWVGYY158 pKa = 9.54ARR160 pKa = 11.84CRR162 pKa = 11.84WMFQNQSEE170 pKa = 4.67ATTHH174 pKa = 4.75LTIYY178 pKa = 10.37EE179 pKa = 4.09FTTRR183 pKa = 11.84RR184 pKa = 11.84DD185 pKa = 3.78SNVGPSLAFQSGLASIQAGVGSNASDD211 pKa = 3.0IGATPFMTPRR221 pKa = 11.84FTEE224 pKa = 3.58NFRR227 pKa = 11.84ILKK230 pKa = 9.67KK231 pKa = 10.87YY232 pKa = 10.38SVEE235 pKa = 3.82LAQGRR240 pKa = 11.84SHH242 pKa = 6.89IHH244 pKa = 4.71TALYY248 pKa = 10.61RR249 pKa = 11.84MNKK252 pKa = 9.61NYY254 pKa = 10.36SDD256 pKa = 4.31SLYY259 pKa = 11.26QMDD262 pKa = 4.21GSSDD266 pKa = 3.62VVLGGWTRR274 pKa = 11.84GLFVIAHH281 pKa = 5.68GTPYY285 pKa = 11.04NSFATKK291 pKa = 10.01TNVSTTPVAIDD302 pKa = 3.12IVANEE307 pKa = 4.4TYY309 pKa = 8.61HH310 pKa = 7.01TYY312 pKa = 11.07TNLQNKK318 pKa = 10.02SIFEE322 pKa = 4.25VQSDD326 pKa = 4.43FPTFTDD332 pKa = 4.18GRR334 pKa = 11.84IIDD337 pKa = 4.27IGSGDD342 pKa = 3.97PEE344 pKa = 4.44AVDD347 pKa = 3.72EE348 pKa = 4.48VV349 pKa = 3.77
MM1 pKa = 7.79RR2 pKa = 11.84RR3 pKa = 11.84RR4 pKa = 11.84PAYY7 pKa = 9.96VGPSWGGSRR16 pKa = 11.84IGGANIPMNTRR27 pKa = 11.84AKK29 pKa = 9.85RR30 pKa = 11.84RR31 pKa = 11.84RR32 pKa = 11.84RR33 pKa = 11.84KK34 pKa = 9.6IIPLLWNSGSKK45 pKa = 9.9LANNLNLGFGSRR57 pKa = 11.84GRR59 pKa = 11.84GPGSMVSRR67 pKa = 11.84IRR69 pKa = 11.84NYY71 pKa = 10.82KK72 pKa = 9.19MEE74 pKa = 4.48ISVKK78 pKa = 9.99PVGTGSSYY86 pKa = 11.02SYY88 pKa = 10.68YY89 pKa = 10.45RR90 pKa = 11.84YY91 pKa = 8.07TRR93 pKa = 11.84RR94 pKa = 11.84PDD96 pKa = 2.87KK97 pKa = 10.71GSRR100 pKa = 11.84IVKK103 pKa = 7.41TQQAPLFSVFNSGSRR118 pKa = 11.84MTSLQGEE125 pKa = 4.25QGYY128 pKa = 6.47TTLSVMTGAKK138 pKa = 9.57LRR140 pKa = 11.84SLYY143 pKa = 11.04LEE145 pKa = 4.34TSANQDD151 pKa = 2.46GRR153 pKa = 11.84FWVGYY158 pKa = 9.54ARR160 pKa = 11.84CRR162 pKa = 11.84WMFQNQSEE170 pKa = 4.67ATTHH174 pKa = 4.75LTIYY178 pKa = 10.37EE179 pKa = 4.09FTTRR183 pKa = 11.84RR184 pKa = 11.84DD185 pKa = 3.78SNVGPSLAFQSGLASIQAGVGSNASDD211 pKa = 3.0IGATPFMTPRR221 pKa = 11.84FTEE224 pKa = 3.58NFRR227 pKa = 11.84ILKK230 pKa = 9.67KK231 pKa = 10.87YY232 pKa = 10.38SVEE235 pKa = 3.82LAQGRR240 pKa = 11.84SHH242 pKa = 6.89IHH244 pKa = 4.71TALYY248 pKa = 10.61RR249 pKa = 11.84MNKK252 pKa = 9.61NYY254 pKa = 10.36SDD256 pKa = 4.31SLYY259 pKa = 11.26QMDD262 pKa = 4.21GSSDD266 pKa = 3.62VVLGGWTRR274 pKa = 11.84GLFVIAHH281 pKa = 5.68GTPYY285 pKa = 11.04NSFATKK291 pKa = 10.01TNVSTTPVAIDD302 pKa = 3.12IVANEE307 pKa = 4.4TYY309 pKa = 8.61HH310 pKa = 7.01TYY312 pKa = 11.07TNLQNKK318 pKa = 10.02SIFEE322 pKa = 4.25VQSDD326 pKa = 4.43FPTFTDD332 pKa = 4.18GRR334 pKa = 11.84IIDD337 pKa = 4.27IGSGDD342 pKa = 3.97PEE344 pKa = 4.44AVDD347 pKa = 3.72EE348 pKa = 4.48VV349 pKa = 3.77
Molecular weight: 38.8 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
655 |
306 |
349 |
327.5 |
37.17 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.107 ± 0.071 | 0.611 ± 0.255 |
5.191 ± 1.155 | 4.58 ± 1.125 |
3.359 ± 0.74 | 7.481 ± 1.781 |
2.443 ± 0.796 | 6.565 ± 0.883 |
5.038 ± 1.035 | 7.023 ± 0.792 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.595 ± 0.213 | 4.427 ± 1.027 |
4.427 ± 0.102 | 4.122 ± 0.313 |
7.786 ± 0.638 | 8.702 ± 1.496 |
7.176 ± 0.893 | 5.344 ± 0.305 |
1.679 ± 0.194 | 5.344 ± 0.372 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
