
Solibacter usitatus (strain Ellin6076)
Taxonomy: cellular organisms; Bacteria; Acidobacteria; Acidobacteriia; Bryobacterales; Solibacteraceae; Candidatus Solibacter; Candidatus Solibacter usitatus
Average proteome isoelectric point is 7.0
Get precalculated fractions of proteins
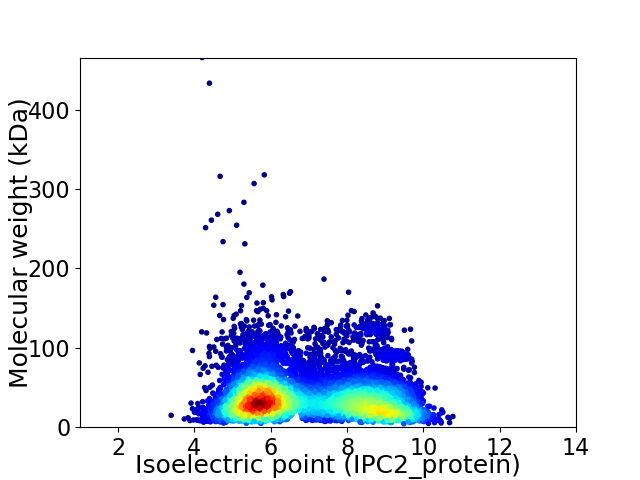
Virtual 2D-PAGE plot for 7761 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|Q01T94|Q01T94_SOLUE Uncharacterized protein OS=Solibacter usitatus (strain Ellin6076) OX=234267 GN=Acid_6200 PE=4 SV=1
MM1 pKa = 7.22SVVDD5 pKa = 4.29RR6 pKa = 11.84EE7 pKa = 4.27FSEE10 pKa = 4.43EE11 pKa = 4.0LTGLMPAVAHH21 pKa = 6.41RR22 pKa = 11.84LGGGACAGYY31 pKa = 9.92IIAVVEE37 pKa = 4.57SNTVEE42 pKa = 4.48LRR44 pKa = 11.84CNLCGEE50 pKa = 4.36IVGVVQVGIMEE61 pKa = 4.62GLLGLDD67 pKa = 4.12CDD69 pKa = 4.77EE70 pKa = 5.07EE71 pKa = 4.46PSPQDD76 pKa = 3.39EE77 pKa = 4.81PEE79 pKa = 4.06DD80 pKa = 3.6
MM1 pKa = 7.22SVVDD5 pKa = 4.29RR6 pKa = 11.84EE7 pKa = 4.27FSEE10 pKa = 4.43EE11 pKa = 4.0LTGLMPAVAHH21 pKa = 6.41RR22 pKa = 11.84LGGGACAGYY31 pKa = 9.92IIAVVEE37 pKa = 4.57SNTVEE42 pKa = 4.48LRR44 pKa = 11.84CNLCGEE50 pKa = 4.36IVGVVQVGIMEE61 pKa = 4.62GLLGLDD67 pKa = 4.12CDD69 pKa = 4.77EE70 pKa = 5.07EE71 pKa = 4.46PSPQDD76 pKa = 3.39EE77 pKa = 4.81PEE79 pKa = 4.06DD80 pKa = 3.6
Molecular weight: 8.43 kDa
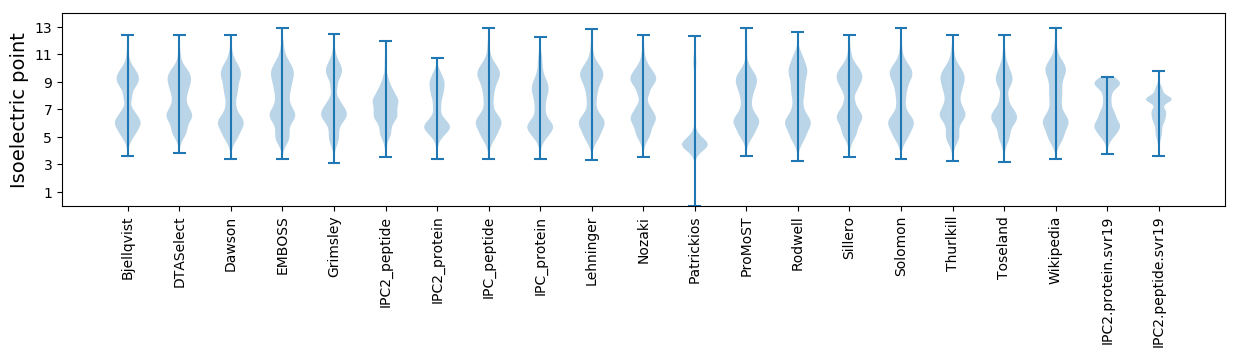
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|Q020U4|Q020U4_SOLUE Tetratricopeptide TPR_2 repeat protein OS=Solibacter usitatus (strain Ellin6076) OX=234267 GN=Acid_3573 PE=4 SV=1
MM1 pKa = 7.35SAKK4 pKa = 10.18SCQLCGKK11 pKa = 8.43PLSRR15 pKa = 11.84LRR17 pKa = 11.84VGGDD21 pKa = 3.38GEE23 pKa = 4.5FCSRR27 pKa = 11.84EE28 pKa = 3.72HH29 pKa = 6.67RR30 pKa = 11.84NQYY33 pKa = 8.89GLRR36 pKa = 11.84RR37 pKa = 11.84GMDD40 pKa = 3.14RR41 pKa = 11.84LQEE44 pKa = 4.0VDD46 pKa = 3.94KK47 pKa = 10.56VTSLMRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84EE56 pKa = 4.18SPRR59 pKa = 11.84QISPARR65 pKa = 11.84LMCNTALATRR75 pKa = 11.84GFLQPRR81 pKa = 11.84TYY83 pKa = 11.06GAGAEE88 pKa = 4.28LAPFAPIFRR97 pKa = 11.84IPEE100 pKa = 3.89PPRR103 pKa = 11.84LSKK106 pKa = 10.58VAEE109 pKa = 4.15RR110 pKa = 11.84YY111 pKa = 7.74VAPRR115 pKa = 11.84TARR118 pKa = 11.84FEE120 pKa = 5.26GIMQVRR126 pKa = 11.84RR127 pKa = 11.84SDD129 pKa = 3.33TRR131 pKa = 11.84QLRR134 pKa = 11.84ITGRR138 pKa = 11.84NSALEE143 pKa = 3.87MPAPKK148 pKa = 9.81RR149 pKa = 11.84RR150 pKa = 11.84LTTQVPQAPMADD162 pKa = 3.76LHH164 pKa = 6.11CTVPAAAASRR174 pKa = 11.84RR175 pKa = 11.84NFALLRR181 pKa = 11.84QTEE184 pKa = 4.25IRR186 pKa = 11.84VHH188 pKa = 7.87AGMPPASLRR197 pKa = 11.84QIALPGTSALGRR209 pKa = 11.84NLPNRR214 pKa = 11.84KK215 pKa = 9.16VGATPLEE222 pKa = 4.58GNALRR227 pKa = 11.84VSIALAFGAPGVSRR241 pKa = 11.84RR242 pKa = 11.84VFVTPPAMSSALVWPRR258 pKa = 11.84TAHH261 pKa = 5.82GVWPEE266 pKa = 4.19TKK268 pKa = 9.3NTTARR273 pKa = 11.84PRR275 pKa = 11.84NLAVRR280 pKa = 11.84MTNPQTCLPAARR292 pKa = 11.84EE293 pKa = 4.2GQRR296 pKa = 11.84RR297 pKa = 11.84SQFVFPGPLRR307 pKa = 11.84PGKK310 pKa = 9.93QKK312 pKa = 10.46PRR314 pKa = 11.84TNGMPPARR322 pKa = 11.84TTDD325 pKa = 3.51VSWILSDD332 pKa = 3.74PRR334 pKa = 11.84TGGIGVPPTSAGFAKK349 pKa = 10.57RR350 pKa = 11.84NGVHH354 pKa = 6.86LCNLTLAPTTTNPTHH369 pKa = 6.75LVAFRR374 pKa = 11.84PFLPQEE380 pKa = 4.04PVGCPMVPFEE390 pKa = 4.48GTTAASIITAPAAGAPGTAADD411 pKa = 4.43APPAAEE417 pKa = 3.84EE418 pKa = 4.07AAPVAQVRR426 pKa = 11.84LEE428 pKa = 3.9EE429 pKa = 4.38HH430 pKa = 6.65FGSGWEE436 pKa = 3.93NWEE439 pKa = 4.11GGTNDD444 pKa = 3.41WLVDD448 pKa = 3.62VAGVRR453 pKa = 11.84TGSLALFQPTMDD465 pKa = 3.63LVDD468 pKa = 3.88YY469 pKa = 10.22EE470 pKa = 5.03LEE472 pKa = 3.81FLARR476 pKa = 11.84IDD478 pKa = 3.82TRR480 pKa = 11.84SLTWVVRR487 pKa = 11.84ATDD490 pKa = 3.41LQEE493 pKa = 4.08YY494 pKa = 7.73MRR496 pKa = 11.84CTLTAIPGGEE506 pKa = 4.24LEE508 pKa = 4.17FSRR511 pKa = 11.84TVVRR515 pKa = 11.84SGIGEE520 pKa = 4.08EE521 pKa = 4.23TVIAPVRR528 pKa = 11.84IPGKK532 pKa = 9.05PRR534 pKa = 11.84AAMTVRR540 pKa = 11.84TRR542 pKa = 11.84VSGDD546 pKa = 3.33TFSVTVDD553 pKa = 3.47GKK555 pKa = 10.86DD556 pKa = 3.01MDD558 pKa = 3.56TWQEE562 pKa = 3.69DD563 pKa = 3.22RR564 pKa = 11.84LYY566 pKa = 10.89CGGIGFMGAHH576 pKa = 7.82DD577 pKa = 4.42DD578 pKa = 3.63RR579 pKa = 11.84ARR581 pKa = 11.84LYY583 pKa = 8.91WVRR586 pKa = 11.84ISSSDD591 pKa = 3.77NIGKK595 pKa = 8.33EE596 pKa = 4.12HH597 pKa = 6.09QKK599 pKa = 10.78KK600 pKa = 10.05
MM1 pKa = 7.35SAKK4 pKa = 10.18SCQLCGKK11 pKa = 8.43PLSRR15 pKa = 11.84LRR17 pKa = 11.84VGGDD21 pKa = 3.38GEE23 pKa = 4.5FCSRR27 pKa = 11.84EE28 pKa = 3.72HH29 pKa = 6.67RR30 pKa = 11.84NQYY33 pKa = 8.89GLRR36 pKa = 11.84RR37 pKa = 11.84GMDD40 pKa = 3.14RR41 pKa = 11.84LQEE44 pKa = 4.0VDD46 pKa = 3.94KK47 pKa = 10.56VTSLMRR53 pKa = 11.84RR54 pKa = 11.84RR55 pKa = 11.84EE56 pKa = 4.18SPRR59 pKa = 11.84QISPARR65 pKa = 11.84LMCNTALATRR75 pKa = 11.84GFLQPRR81 pKa = 11.84TYY83 pKa = 11.06GAGAEE88 pKa = 4.28LAPFAPIFRR97 pKa = 11.84IPEE100 pKa = 3.89PPRR103 pKa = 11.84LSKK106 pKa = 10.58VAEE109 pKa = 4.15RR110 pKa = 11.84YY111 pKa = 7.74VAPRR115 pKa = 11.84TARR118 pKa = 11.84FEE120 pKa = 5.26GIMQVRR126 pKa = 11.84RR127 pKa = 11.84SDD129 pKa = 3.33TRR131 pKa = 11.84QLRR134 pKa = 11.84ITGRR138 pKa = 11.84NSALEE143 pKa = 3.87MPAPKK148 pKa = 9.81RR149 pKa = 11.84RR150 pKa = 11.84LTTQVPQAPMADD162 pKa = 3.76LHH164 pKa = 6.11CTVPAAAASRR174 pKa = 11.84RR175 pKa = 11.84NFALLRR181 pKa = 11.84QTEE184 pKa = 4.25IRR186 pKa = 11.84VHH188 pKa = 7.87AGMPPASLRR197 pKa = 11.84QIALPGTSALGRR209 pKa = 11.84NLPNRR214 pKa = 11.84KK215 pKa = 9.16VGATPLEE222 pKa = 4.58GNALRR227 pKa = 11.84VSIALAFGAPGVSRR241 pKa = 11.84RR242 pKa = 11.84VFVTPPAMSSALVWPRR258 pKa = 11.84TAHH261 pKa = 5.82GVWPEE266 pKa = 4.19TKK268 pKa = 9.3NTTARR273 pKa = 11.84PRR275 pKa = 11.84NLAVRR280 pKa = 11.84MTNPQTCLPAARR292 pKa = 11.84EE293 pKa = 4.2GQRR296 pKa = 11.84RR297 pKa = 11.84SQFVFPGPLRR307 pKa = 11.84PGKK310 pKa = 9.93QKK312 pKa = 10.46PRR314 pKa = 11.84TNGMPPARR322 pKa = 11.84TTDD325 pKa = 3.51VSWILSDD332 pKa = 3.74PRR334 pKa = 11.84TGGIGVPPTSAGFAKK349 pKa = 10.57RR350 pKa = 11.84NGVHH354 pKa = 6.86LCNLTLAPTTTNPTHH369 pKa = 6.75LVAFRR374 pKa = 11.84PFLPQEE380 pKa = 4.04PVGCPMVPFEE390 pKa = 4.48GTTAASIITAPAAGAPGTAADD411 pKa = 4.43APPAAEE417 pKa = 3.84EE418 pKa = 4.07AAPVAQVRR426 pKa = 11.84LEE428 pKa = 3.9EE429 pKa = 4.38HH430 pKa = 6.65FGSGWEE436 pKa = 3.93NWEE439 pKa = 4.11GGTNDD444 pKa = 3.41WLVDD448 pKa = 3.62VAGVRR453 pKa = 11.84TGSLALFQPTMDD465 pKa = 3.63LVDD468 pKa = 3.88YY469 pKa = 10.22EE470 pKa = 5.03LEE472 pKa = 3.81FLARR476 pKa = 11.84IDD478 pKa = 3.82TRR480 pKa = 11.84SLTWVVRR487 pKa = 11.84ATDD490 pKa = 3.41LQEE493 pKa = 4.08YY494 pKa = 7.73MRR496 pKa = 11.84CTLTAIPGGEE506 pKa = 4.24LEE508 pKa = 4.17FSRR511 pKa = 11.84TVVRR515 pKa = 11.84SGIGEE520 pKa = 4.08EE521 pKa = 4.23TVIAPVRR528 pKa = 11.84IPGKK532 pKa = 9.05PRR534 pKa = 11.84AAMTVRR540 pKa = 11.84TRR542 pKa = 11.84VSGDD546 pKa = 3.33TFSVTVDD553 pKa = 3.47GKK555 pKa = 10.86DD556 pKa = 3.01MDD558 pKa = 3.56TWQEE562 pKa = 3.69DD563 pKa = 3.22RR564 pKa = 11.84LYY566 pKa = 10.89CGGIGFMGAHH576 pKa = 7.82DD577 pKa = 4.42DD578 pKa = 3.63RR579 pKa = 11.84ARR581 pKa = 11.84LYY583 pKa = 8.91WVRR586 pKa = 11.84ISSSDD591 pKa = 3.77NIGKK595 pKa = 8.33EE596 pKa = 4.12HH597 pKa = 6.09QKK599 pKa = 10.78KK600 pKa = 10.05
Molecular weight: 65.49 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
2980018 |
37 |
4666 |
384.0 |
41.85 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
11.392 ± 0.033 | 0.899 ± 0.008 |
4.985 ± 0.023 | 5.217 ± 0.035 |
3.995 ± 0.018 | 8.555 ± 0.029 |
2.0 ± 0.013 | 4.791 ± 0.017 |
3.451 ± 0.026 | 9.813 ± 0.031 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.227 ± 0.012 | 3.35 ± 0.028 |
5.591 ± 0.022 | 3.562 ± 0.016 |
6.834 ± 0.032 | 5.977 ± 0.031 |
5.796 ± 0.041 | 7.324 ± 0.023 |
1.474 ± 0.012 | 2.766 ± 0.015 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
