
Shuangao Bedbug Virus 2
Taxonomy: Viruses; Riboviria; unclassified Riboviria; unclassified ssRNA viruses; unclassified ssRNA negative-strand viruses
Average proteome isoelectric point is 7.95
Get precalculated fractions of proteins
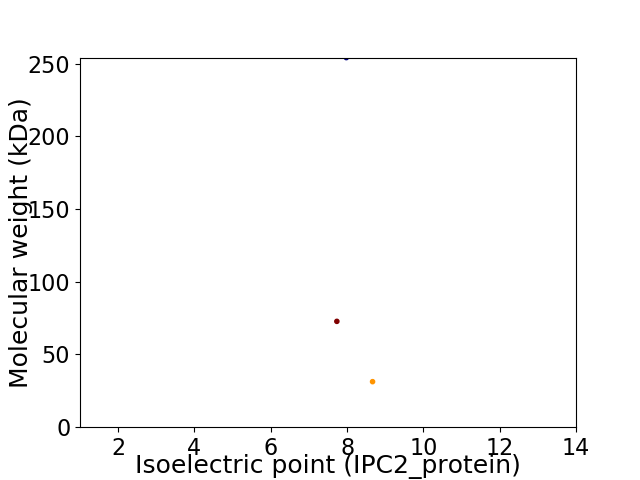
Virtual 2D-PAGE plot for 3 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0B5KXF4|A0A0B5KXF4_9VIRU Glycoprotein OS=Shuangao Bedbug Virus 2 OX=1608072 GN=G PE=4 SV=1
MM1 pKa = 7.29CQFPNMLMFILILKK15 pKa = 10.2SGVTGQHH22 pKa = 6.47PKK24 pKa = 10.77LSFISHH30 pKa = 6.95LDD32 pKa = 2.81KK33 pKa = 11.78GEE35 pKa = 3.84AVKK38 pKa = 10.59INPIPQCKK46 pKa = 8.74TDD48 pKa = 3.08WSKK51 pKa = 8.04TTVYY55 pKa = 10.8LPGTLMVLRR64 pKa = 11.84PNPKK68 pKa = 9.04LLKK71 pKa = 10.61SIGLLATAFTVKK83 pKa = 9.75TKK85 pKa = 10.68CEE87 pKa = 3.8KK88 pKa = 10.6FLFSQNVRR96 pKa = 11.84TTIYY100 pKa = 10.37DD101 pKa = 3.54SPIPWSSFDD110 pKa = 3.62DD111 pKa = 3.61SFIRR115 pKa = 11.84FFEE118 pKa = 4.13EE119 pKa = 5.4KK120 pKa = 8.68YY121 pKa = 8.81TKK123 pKa = 9.99PVQLEE128 pKa = 3.93KK129 pKa = 11.04LQLKK133 pKa = 10.45VSGEE137 pKa = 3.95EE138 pKa = 4.95LMGQDD143 pKa = 5.19LYY145 pKa = 11.93SCGYY149 pKa = 10.49YY150 pKa = 10.17SGQFQNINYY159 pKa = 9.1IKK161 pKa = 9.93FKK163 pKa = 10.7AVEE166 pKa = 4.4TEE168 pKa = 3.43WDD170 pKa = 3.72FQGNLLSPIFDD181 pKa = 4.07PSCLHH186 pKa = 5.85NHH188 pKa = 5.96TSCVLKK194 pKa = 10.45EE195 pKa = 3.88DD196 pKa = 3.76SKK198 pKa = 11.8LYY200 pKa = 10.83FPMGSPNLGHH210 pKa = 6.88CEE212 pKa = 3.59FSTISTLAGFFLSQSSKK229 pKa = 8.95ISSFVSHH236 pKa = 6.62NSAFKK241 pKa = 10.29TYY243 pKa = 10.56FDD245 pKa = 3.74WSLKK249 pKa = 9.52GSHH252 pKa = 6.7PRR254 pKa = 11.84SLCHH258 pKa = 6.57YY259 pKa = 8.53ITGNHH264 pKa = 6.41YY265 pKa = 8.74ITLEE269 pKa = 4.04GHH271 pKa = 5.37VVRR274 pKa = 11.84ALDD277 pKa = 3.74SEE279 pKa = 4.24GRR281 pKa = 11.84DD282 pKa = 3.23IFIKK286 pKa = 10.39SQPNPLFSQPGNHH299 pKa = 6.56RR300 pKa = 11.84IKK302 pKa = 10.58RR303 pKa = 11.84SEE305 pKa = 4.18HH306 pKa = 6.35LPLTTINTTLSQLEE320 pKa = 4.41FPKK323 pKa = 10.6FPPPSLKK330 pKa = 10.13FRR332 pKa = 11.84TSQSVSRR339 pKa = 11.84SKK341 pKa = 10.78RR342 pKa = 11.84SYY344 pKa = 10.9NYY346 pKa = 8.66YY347 pKa = 10.45NYY349 pKa = 9.29PARR352 pKa = 11.84QRR354 pKa = 11.84EE355 pKa = 4.03QMLYY359 pKa = 9.89EE360 pKa = 4.17HH361 pKa = 6.95SSWTLSEE368 pKa = 5.1LEE370 pKa = 4.37FEE372 pKa = 5.65LSLLHH377 pKa = 6.39NEE379 pKa = 4.82SIAEE383 pKa = 3.98IQKK386 pKa = 10.34RR387 pKa = 11.84WILEE391 pKa = 4.15CKK393 pKa = 9.96LKK395 pKa = 10.72NLHH398 pKa = 5.85GQLARR403 pKa = 11.84SVLHH407 pKa = 6.6IDD409 pKa = 3.08PRR411 pKa = 11.84PYY413 pKa = 9.51IAYY416 pKa = 9.52VLSSQAFITLNVNSKK431 pKa = 8.89TYY433 pKa = 10.16YY434 pKa = 9.97ISGDD438 pKa = 3.9PVWDD442 pKa = 3.29IEE444 pKa = 4.5IYY446 pKa = 10.43KK447 pKa = 9.6PYY449 pKa = 9.84KK450 pKa = 7.88WCNGSLYY457 pKa = 10.75VHH459 pKa = 6.13TEE461 pKa = 3.91KK462 pKa = 11.26GPGWLKK468 pKa = 10.91NEE470 pKa = 3.77WGIVISGDD478 pKa = 3.88LLTGGCNGSLYY489 pKa = 10.84VHH491 pKa = 6.13TEE493 pKa = 3.91KK494 pKa = 11.26GPGWLKK500 pKa = 10.91NEE502 pKa = 3.77WGIVISGDD510 pKa = 3.87LLTGGCTSNQAPEE523 pKa = 4.15IFLFPTMRR531 pKa = 11.84NGSWDD536 pKa = 4.13LISNRR541 pKa = 11.84SLSSDD546 pKa = 3.34LSLPSPHH553 pKa = 6.94HH554 pKa = 5.8LLEE557 pKa = 5.15HH558 pKa = 6.49GFYY561 pKa = 9.01KK562 pKa = 10.49TSFEE566 pKa = 4.5IDD568 pKa = 2.92TSDD571 pKa = 3.2ALLRR575 pKa = 11.84SLEE578 pKa = 4.37AGLSSMSYY586 pKa = 9.59GASSRR591 pKa = 11.84KK592 pKa = 9.2SAGVSLWKK600 pKa = 10.5IPDD603 pKa = 4.25PPSSWYY609 pKa = 10.43EE610 pKa = 3.73SMILHH615 pKa = 6.99FLTPTTMLIFGLIIFLFIWRR635 pKa = 11.84FLRR638 pKa = 11.84GG639 pKa = 3.21
MM1 pKa = 7.29CQFPNMLMFILILKK15 pKa = 10.2SGVTGQHH22 pKa = 6.47PKK24 pKa = 10.77LSFISHH30 pKa = 6.95LDD32 pKa = 2.81KK33 pKa = 11.78GEE35 pKa = 3.84AVKK38 pKa = 10.59INPIPQCKK46 pKa = 8.74TDD48 pKa = 3.08WSKK51 pKa = 8.04TTVYY55 pKa = 10.8LPGTLMVLRR64 pKa = 11.84PNPKK68 pKa = 9.04LLKK71 pKa = 10.61SIGLLATAFTVKK83 pKa = 9.75TKK85 pKa = 10.68CEE87 pKa = 3.8KK88 pKa = 10.6FLFSQNVRR96 pKa = 11.84TTIYY100 pKa = 10.37DD101 pKa = 3.54SPIPWSSFDD110 pKa = 3.62DD111 pKa = 3.61SFIRR115 pKa = 11.84FFEE118 pKa = 4.13EE119 pKa = 5.4KK120 pKa = 8.68YY121 pKa = 8.81TKK123 pKa = 9.99PVQLEE128 pKa = 3.93KK129 pKa = 11.04LQLKK133 pKa = 10.45VSGEE137 pKa = 3.95EE138 pKa = 4.95LMGQDD143 pKa = 5.19LYY145 pKa = 11.93SCGYY149 pKa = 10.49YY150 pKa = 10.17SGQFQNINYY159 pKa = 9.1IKK161 pKa = 9.93FKK163 pKa = 10.7AVEE166 pKa = 4.4TEE168 pKa = 3.43WDD170 pKa = 3.72FQGNLLSPIFDD181 pKa = 4.07PSCLHH186 pKa = 5.85NHH188 pKa = 5.96TSCVLKK194 pKa = 10.45EE195 pKa = 3.88DD196 pKa = 3.76SKK198 pKa = 11.8LYY200 pKa = 10.83FPMGSPNLGHH210 pKa = 6.88CEE212 pKa = 3.59FSTISTLAGFFLSQSSKK229 pKa = 8.95ISSFVSHH236 pKa = 6.62NSAFKK241 pKa = 10.29TYY243 pKa = 10.56FDD245 pKa = 3.74WSLKK249 pKa = 9.52GSHH252 pKa = 6.7PRR254 pKa = 11.84SLCHH258 pKa = 6.57YY259 pKa = 8.53ITGNHH264 pKa = 6.41YY265 pKa = 8.74ITLEE269 pKa = 4.04GHH271 pKa = 5.37VVRR274 pKa = 11.84ALDD277 pKa = 3.74SEE279 pKa = 4.24GRR281 pKa = 11.84DD282 pKa = 3.23IFIKK286 pKa = 10.39SQPNPLFSQPGNHH299 pKa = 6.56RR300 pKa = 11.84IKK302 pKa = 10.58RR303 pKa = 11.84SEE305 pKa = 4.18HH306 pKa = 6.35LPLTTINTTLSQLEE320 pKa = 4.41FPKK323 pKa = 10.6FPPPSLKK330 pKa = 10.13FRR332 pKa = 11.84TSQSVSRR339 pKa = 11.84SKK341 pKa = 10.78RR342 pKa = 11.84SYY344 pKa = 10.9NYY346 pKa = 8.66YY347 pKa = 10.45NYY349 pKa = 9.29PARR352 pKa = 11.84QRR354 pKa = 11.84EE355 pKa = 4.03QMLYY359 pKa = 9.89EE360 pKa = 4.17HH361 pKa = 6.95SSWTLSEE368 pKa = 5.1LEE370 pKa = 4.37FEE372 pKa = 5.65LSLLHH377 pKa = 6.39NEE379 pKa = 4.82SIAEE383 pKa = 3.98IQKK386 pKa = 10.34RR387 pKa = 11.84WILEE391 pKa = 4.15CKK393 pKa = 9.96LKK395 pKa = 10.72NLHH398 pKa = 5.85GQLARR403 pKa = 11.84SVLHH407 pKa = 6.6IDD409 pKa = 3.08PRR411 pKa = 11.84PYY413 pKa = 9.51IAYY416 pKa = 9.52VLSSQAFITLNVNSKK431 pKa = 8.89TYY433 pKa = 10.16YY434 pKa = 9.97ISGDD438 pKa = 3.9PVWDD442 pKa = 3.29IEE444 pKa = 4.5IYY446 pKa = 10.43KK447 pKa = 9.6PYY449 pKa = 9.84KK450 pKa = 7.88WCNGSLYY457 pKa = 10.75VHH459 pKa = 6.13TEE461 pKa = 3.91KK462 pKa = 11.26GPGWLKK468 pKa = 10.91NEE470 pKa = 3.77WGIVISGDD478 pKa = 3.88LLTGGCNGSLYY489 pKa = 10.84VHH491 pKa = 6.13TEE493 pKa = 3.91KK494 pKa = 11.26GPGWLKK500 pKa = 10.91NEE502 pKa = 3.77WGIVISGDD510 pKa = 3.87LLTGGCTSNQAPEE523 pKa = 4.15IFLFPTMRR531 pKa = 11.84NGSWDD536 pKa = 4.13LISNRR541 pKa = 11.84SLSSDD546 pKa = 3.34LSLPSPHH553 pKa = 6.94HH554 pKa = 5.8LLEE557 pKa = 5.15HH558 pKa = 6.49GFYY561 pKa = 9.01KK562 pKa = 10.49TSFEE566 pKa = 4.5IDD568 pKa = 2.92TSDD571 pKa = 3.2ALLRR575 pKa = 11.84SLEE578 pKa = 4.37AGLSSMSYY586 pKa = 9.59GASSRR591 pKa = 11.84KK592 pKa = 9.2SAGVSLWKK600 pKa = 10.5IPDD603 pKa = 4.25PPSSWYY609 pKa = 10.43EE610 pKa = 3.73SMILHH615 pKa = 6.99FLTPTTMLIFGLIIFLFIWRR635 pKa = 11.84FLRR638 pKa = 11.84GG639 pKa = 3.21
Molecular weight: 72.79 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0B5KXF4|A0A0B5KXF4_9VIRU Glycoprotein OS=Shuangao Bedbug Virus 2 OX=1608072 GN=G PE=4 SV=1
MM1 pKa = 7.73AEE3 pKa = 4.36SSLSTTQNHH12 pKa = 7.45KK13 pKa = 10.54ILLPKK18 pKa = 9.87NASSLTITPVEE29 pKa = 4.14QFPHH33 pKa = 8.07LDD35 pKa = 2.91IQRR38 pKa = 11.84YY39 pKa = 7.97PEE41 pKa = 4.16QKK43 pKa = 9.45SSHH46 pKa = 6.64HH47 pKa = 5.72IVIGVGNQEE56 pKa = 4.12AVWRR60 pKa = 11.84TEE62 pKa = 3.85KK63 pKa = 10.73EE64 pKa = 4.08VLSPVILHH72 pKa = 6.66LEE74 pKa = 4.32SACTVANANIPVRR87 pKa = 11.84RR88 pKa = 11.84TFKK91 pKa = 10.89LVLGLALLDD100 pKa = 3.87IYY102 pKa = 11.2HH103 pKa = 6.7NSGVSNLTIQKK114 pKa = 9.55IFSSCLEE121 pKa = 3.97IMKK124 pKa = 10.21KK125 pKa = 7.41EE126 pKa = 3.64QWRR129 pKa = 11.84IYY131 pKa = 9.09EE132 pKa = 4.51DD133 pKa = 3.1KK134 pKa = 9.92HH135 pKa = 5.69TSSVAWKK142 pKa = 9.78FLFKK146 pKa = 10.59LISVTPCIPSNVVNVKK162 pKa = 9.86NKK164 pKa = 8.52KK165 pKa = 7.27TFSGVISNGPYY176 pKa = 8.37SGLYY180 pKa = 9.5ISHH183 pKa = 6.95TFNYY187 pKa = 9.58HH188 pKa = 3.98SQEE191 pKa = 3.71YY192 pKa = 10.46DD193 pKa = 3.28LAEE196 pKa = 4.57SIYY199 pKa = 10.31HH200 pKa = 6.55RR201 pKa = 11.84SSGHH205 pKa = 5.66YY206 pKa = 9.86IIEE209 pKa = 3.98QAPNDD214 pKa = 3.95GKK216 pKa = 9.72EE217 pKa = 4.05TFIWMWDD224 pKa = 3.35QTRR227 pKa = 11.84TLPQEE232 pKa = 3.54FHH234 pKa = 6.54ARR236 pKa = 11.84LSIMEE241 pKa = 4.44HH242 pKa = 4.82YY243 pKa = 10.34KK244 pKa = 10.56KK245 pKa = 10.87YY246 pKa = 11.02NHH248 pKa = 6.75TKK250 pKa = 9.74TKK252 pKa = 10.74AITNKK257 pKa = 9.86KK258 pKa = 7.67SQEE261 pKa = 3.74YY262 pKa = 9.44DD263 pKa = 3.42APWNKK268 pKa = 10.23KK269 pKa = 6.8KK270 pKa = 9.33TKK272 pKa = 10.18
MM1 pKa = 7.73AEE3 pKa = 4.36SSLSTTQNHH12 pKa = 7.45KK13 pKa = 10.54ILLPKK18 pKa = 9.87NASSLTITPVEE29 pKa = 4.14QFPHH33 pKa = 8.07LDD35 pKa = 2.91IQRR38 pKa = 11.84YY39 pKa = 7.97PEE41 pKa = 4.16QKK43 pKa = 9.45SSHH46 pKa = 6.64HH47 pKa = 5.72IVIGVGNQEE56 pKa = 4.12AVWRR60 pKa = 11.84TEE62 pKa = 3.85KK63 pKa = 10.73EE64 pKa = 4.08VLSPVILHH72 pKa = 6.66LEE74 pKa = 4.32SACTVANANIPVRR87 pKa = 11.84RR88 pKa = 11.84TFKK91 pKa = 10.89LVLGLALLDD100 pKa = 3.87IYY102 pKa = 11.2HH103 pKa = 6.7NSGVSNLTIQKK114 pKa = 9.55IFSSCLEE121 pKa = 3.97IMKK124 pKa = 10.21KK125 pKa = 7.41EE126 pKa = 3.64QWRR129 pKa = 11.84IYY131 pKa = 9.09EE132 pKa = 4.51DD133 pKa = 3.1KK134 pKa = 9.92HH135 pKa = 5.69TSSVAWKK142 pKa = 9.78FLFKK146 pKa = 10.59LISVTPCIPSNVVNVKK162 pKa = 9.86NKK164 pKa = 8.52KK165 pKa = 7.27TFSGVISNGPYY176 pKa = 8.37SGLYY180 pKa = 9.5ISHH183 pKa = 6.95TFNYY187 pKa = 9.58HH188 pKa = 3.98SQEE191 pKa = 3.71YY192 pKa = 10.46DD193 pKa = 3.28LAEE196 pKa = 4.57SIYY199 pKa = 10.31HH200 pKa = 6.55RR201 pKa = 11.84SSGHH205 pKa = 5.66YY206 pKa = 9.86IIEE209 pKa = 3.98QAPNDD214 pKa = 3.95GKK216 pKa = 9.72EE217 pKa = 4.05TFIWMWDD224 pKa = 3.35QTRR227 pKa = 11.84TLPQEE232 pKa = 3.54FHH234 pKa = 6.54ARR236 pKa = 11.84LSIMEE241 pKa = 4.44HH242 pKa = 4.82YY243 pKa = 10.34KK244 pKa = 10.56KK245 pKa = 10.87YY246 pKa = 11.02NHH248 pKa = 6.75TKK250 pKa = 9.74TKK252 pKa = 10.74AITNKK257 pKa = 9.86KK258 pKa = 7.67SQEE261 pKa = 3.74YY262 pKa = 9.44DD263 pKa = 3.42APWNKK268 pKa = 10.23KK269 pKa = 6.8KK270 pKa = 9.33TKK272 pKa = 10.18
Molecular weight: 31.32 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
3118 |
272 |
2207 |
1039.3 |
119.43 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
3.849 ± 0.416 | 1.86 ± 0.196 |
3.784 ± 0.392 | 5.099 ± 0.314 |
5.388 ± 0.501 | 4.201 ± 0.779 |
3.72 ± 0.343 | 8.146 ± 0.61 |
6.414 ± 0.706 | 11.482 ± 0.951 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.213 ± 0.324 | 5.484 ± 0.449 |
5.516 ± 0.266 | 3.624 ± 0.192 |
4.009 ± 0.422 | 9.397 ± 1.025 |
5.901 ± 0.269 | 4.201 ± 0.402 |
1.668 ± 0.402 | 4.041 ± 0.143 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
