
Nakamurella silvestris
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Actinobacteria; Actinomycetia; Nakamurellales; Nakamurellaceae; Nakamurella
Average proteome isoelectric point is 6.11
Get precalculated fractions of proteins
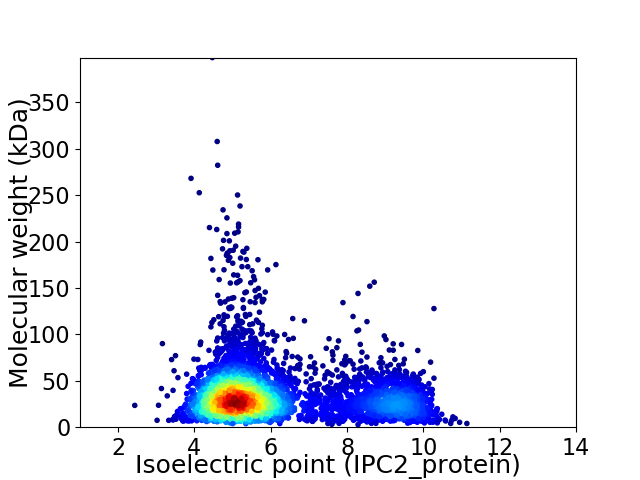
Virtual 2D-PAGE plot for 4524 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A399VQY1|A0A399VQY1_9ACTN Nucleotide-binding protein D1871_00570 OS=Nakamurella silvestris OX=1645681 GN=rapZ PE=3 SV=1
MM1 pKa = 7.83SDD3 pKa = 2.92RR4 pKa = 11.84TVTFNHH10 pKa = 7.15LGNCIINADD19 pKa = 3.59QTGTDD24 pKa = 4.07DD25 pKa = 4.13YY26 pKa = 11.65VAASTKK32 pKa = 7.24TQQIAVGAAAQAISITTPLPDD53 pKa = 3.79APVVGDD59 pKa = 4.49ALDD62 pKa = 3.77LAATGGASGNVVTFSTTTTGICTVTGTTVTFDD94 pKa = 3.38HH95 pKa = 7.21VGDD98 pKa = 4.13CLINADD104 pKa = 3.71QTGTTDD110 pKa = 3.24YY111 pKa = 9.91TAAPTATRR119 pKa = 11.84SVTIGQAPQAISFTTTEE136 pKa = 4.08PDD138 pKa = 3.27TAVVGDD144 pKa = 4.33SYY146 pKa = 11.67EE147 pKa = 4.01VAAVGGALRR156 pKa = 11.84SNPVNRR162 pKa = 11.84SLAADD167 pKa = 4.17GSGEE171 pKa = 4.12PVTFSTTSPEE181 pKa = 3.54ICTVSGSTVTFDD193 pKa = 4.39HH194 pKa = 7.34VGDD197 pKa = 4.13CLINADD203 pKa = 3.86QAGDD207 pKa = 3.95DD208 pKa = 5.11DD209 pKa = 4.46YY210 pKa = 11.59TAAPTIIQTVTVGQAPQAISFTTTEE235 pKa = 3.81PEE237 pKa = 4.02APVVGQTHH245 pKa = 5.61TVAATGGPSGNEE257 pKa = 3.85VTFSTPSAEE266 pKa = 4.03VCTVNGATVTFTHH279 pKa = 7.16PGDD282 pKa = 3.92CVVNADD288 pKa = 3.88QVGNDD293 pKa = 4.8DD294 pKa = 3.77YY295 pKa = 11.39TAAPTTTQTLTIGKK309 pKa = 9.43APQTVEE315 pKa = 4.39FSSTPPSDD323 pKa = 3.89AKK325 pKa = 11.14AGDD328 pKa = 3.6TYY330 pKa = 11.51EE331 pKa = 4.2PVATGGTSGTPVVFSIDD348 pKa = 3.5PAGTPGACEE357 pKa = 3.43IVAGKK362 pKa = 10.35VLFTGPGTCIITAEE376 pKa = 4.03QAGDD380 pKa = 3.9ADD382 pKa = 4.31HH383 pKa = 7.15LPTTSQQTVTVAAGEE398 pKa = 4.25VTITTEE404 pKa = 4.14ALPHH408 pKa = 5.98GAVGLDD414 pKa = 3.42YY415 pKa = 7.66TTSVQAEE422 pKa = 4.45GGLAPYY428 pKa = 9.91AWAVAAGSLPDD439 pKa = 4.89GLALDD444 pKa = 4.96PDD446 pKa = 4.2SGDD449 pKa = 3.05ITGTPTQPGAFTFTISATDD468 pKa = 3.72SEE470 pKa = 4.54NQPVAGTRR478 pKa = 11.84AFTLTVLAEE487 pKa = 4.24EE488 pKa = 4.31VLPPVVDD495 pKa = 4.22PPTGPTGEE503 pKa = 4.3PTTTPVPTTGSTTAVTPGTTTPGAPGTTPPPPTALPATGVNTLPLILLGLVLLVAGLLLNSRR565 pKa = 11.84IGRR568 pKa = 11.84NRR570 pKa = 11.84HH571 pKa = 3.37QHH573 pKa = 4.58
MM1 pKa = 7.83SDD3 pKa = 2.92RR4 pKa = 11.84TVTFNHH10 pKa = 7.15LGNCIINADD19 pKa = 3.59QTGTDD24 pKa = 4.07DD25 pKa = 4.13YY26 pKa = 11.65VAASTKK32 pKa = 7.24TQQIAVGAAAQAISITTPLPDD53 pKa = 3.79APVVGDD59 pKa = 4.49ALDD62 pKa = 3.77LAATGGASGNVVTFSTTTTGICTVTGTTVTFDD94 pKa = 3.38HH95 pKa = 7.21VGDD98 pKa = 4.13CLINADD104 pKa = 3.71QTGTTDD110 pKa = 3.24YY111 pKa = 9.91TAAPTATRR119 pKa = 11.84SVTIGQAPQAISFTTTEE136 pKa = 4.08PDD138 pKa = 3.27TAVVGDD144 pKa = 4.33SYY146 pKa = 11.67EE147 pKa = 4.01VAAVGGALRR156 pKa = 11.84SNPVNRR162 pKa = 11.84SLAADD167 pKa = 4.17GSGEE171 pKa = 4.12PVTFSTTSPEE181 pKa = 3.54ICTVSGSTVTFDD193 pKa = 4.39HH194 pKa = 7.34VGDD197 pKa = 4.13CLINADD203 pKa = 3.86QAGDD207 pKa = 3.95DD208 pKa = 5.11DD209 pKa = 4.46YY210 pKa = 11.59TAAPTIIQTVTVGQAPQAISFTTTEE235 pKa = 3.81PEE237 pKa = 4.02APVVGQTHH245 pKa = 5.61TVAATGGPSGNEE257 pKa = 3.85VTFSTPSAEE266 pKa = 4.03VCTVNGATVTFTHH279 pKa = 7.16PGDD282 pKa = 3.92CVVNADD288 pKa = 3.88QVGNDD293 pKa = 4.8DD294 pKa = 3.77YY295 pKa = 11.39TAAPTTTQTLTIGKK309 pKa = 9.43APQTVEE315 pKa = 4.39FSSTPPSDD323 pKa = 3.89AKK325 pKa = 11.14AGDD328 pKa = 3.6TYY330 pKa = 11.51EE331 pKa = 4.2PVATGGTSGTPVVFSIDD348 pKa = 3.5PAGTPGACEE357 pKa = 3.43IVAGKK362 pKa = 10.35VLFTGPGTCIITAEE376 pKa = 4.03QAGDD380 pKa = 3.9ADD382 pKa = 4.31HH383 pKa = 7.15LPTTSQQTVTVAAGEE398 pKa = 4.25VTITTEE404 pKa = 4.14ALPHH408 pKa = 5.98GAVGLDD414 pKa = 3.42YY415 pKa = 7.66TTSVQAEE422 pKa = 4.45GGLAPYY428 pKa = 9.91AWAVAAGSLPDD439 pKa = 4.89GLALDD444 pKa = 4.96PDD446 pKa = 4.2SGDD449 pKa = 3.05ITGTPTQPGAFTFTISATDD468 pKa = 3.72SEE470 pKa = 4.54NQPVAGTRR478 pKa = 11.84AFTLTVLAEE487 pKa = 4.24EE488 pKa = 4.31VLPPVVDD495 pKa = 4.22PPTGPTGEE503 pKa = 4.3PTTTPVPTTGSTTAVTPGTTTPGAPGTTPPPPTALPATGVNTLPLILLGLVLLVAGLLLNSRR565 pKa = 11.84IGRR568 pKa = 11.84NRR570 pKa = 11.84HH571 pKa = 3.37QHH573 pKa = 4.58
Molecular weight: 57.15 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A399UPE8|A0A399UPE8_9ACTN Beta-N-acetylhexosaminidase OS=Nakamurella silvestris OX=1645681 GN=D1871_21610 PE=3 SV=1
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.7LLRR22 pKa = 11.84KK23 pKa = 7.81TRR25 pKa = 11.84VARR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.68AGKK33 pKa = 9.73
MM1 pKa = 7.4GSVIKK6 pKa = 10.42KK7 pKa = 8.47RR8 pKa = 11.84RR9 pKa = 11.84KK10 pKa = 9.22RR11 pKa = 11.84MAKK14 pKa = 9.41KK15 pKa = 9.87KK16 pKa = 9.7HH17 pKa = 5.84RR18 pKa = 11.84KK19 pKa = 8.7LLRR22 pKa = 11.84KK23 pKa = 7.81TRR25 pKa = 11.84VARR28 pKa = 11.84RR29 pKa = 11.84KK30 pKa = 9.68AGKK33 pKa = 9.73
Molecular weight: 4.0 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1536067 |
27 |
3932 |
339.5 |
36.11 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
12.827 ± 0.052 | 0.701 ± 0.011 |
5.934 ± 0.029 | 5.007 ± 0.04 |
2.954 ± 0.02 | 9.353 ± 0.035 |
1.942 ± 0.018 | 4.299 ± 0.025 |
2.327 ± 0.026 | 9.72 ± 0.057 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.725 ± 0.017 | 2.367 ± 0.035 |
5.787 ± 0.034 | 2.99 ± 0.018 |
6.623 ± 0.052 | 6.051 ± 0.03 |
7.068 ± 0.074 | 8.826 ± 0.035 |
1.433 ± 0.015 | 2.065 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
