
Brugia malayi (Filarial nematode worm)
Taxonomy: cellular organisms; Eukaryota; Opisthokonta; Metazoa; Eumetazoa; Bilateria; Protostomia; Ecdysozoa; Nematoda; Chromadorea; Rhabditida; Spirurina; Spiruromorpha; Filarioidea; Onchocercidae; Brugia
Average proteome isoelectric point is 6.84
Get precalculated fractions of proteins
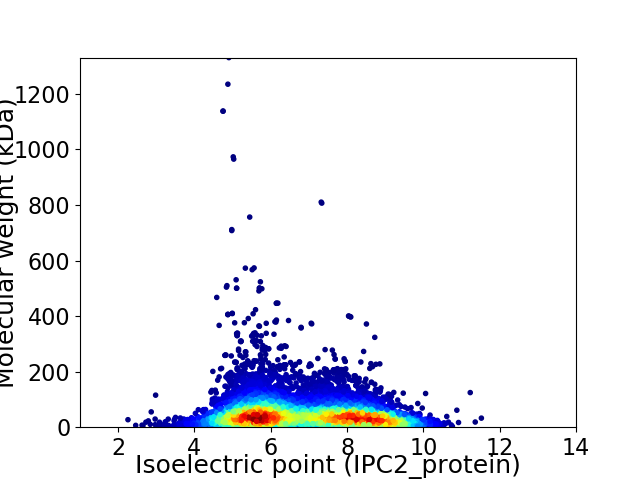
Virtual 2D-PAGE plot for 10206 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A5S6PD84|A0A5S6PD84_BRUMA Uncharacterized protein OS=Brugia malayi OX=6279 PE=4 SV=1
MM1 pKa = 7.14VSSDD5 pKa = 3.78EE6 pKa = 4.89FRR8 pKa = 11.84DD9 pKa = 3.57ADD11 pKa = 3.74DD12 pKa = 4.2VLFSTSNEE20 pKa = 3.63VTYY23 pKa = 10.59IAFDD27 pKa = 3.82GADD30 pKa = 3.43CKK32 pKa = 10.11YY33 pKa = 9.69TPGDD37 pKa = 3.82CFDD40 pKa = 4.45PAVMTRR46 pKa = 11.84IKK48 pKa = 10.42LSCHH52 pKa = 5.29SRR54 pKa = 11.84LFNATSFDD62 pKa = 4.25QIPEE66 pKa = 4.23LLDD69 pKa = 5.09DD70 pKa = 5.68IIIPDD75 pKa = 3.74YY76 pKa = 11.17CSFGDD81 pKa = 4.56SVDD84 pKa = 3.22INIWIGPSEE93 pKa = 4.39NVSSPYY99 pKa = 10.03FDD101 pKa = 4.0PKK103 pKa = 11.44SNIFAKK109 pKa = 10.65
MM1 pKa = 7.14VSSDD5 pKa = 3.78EE6 pKa = 4.89FRR8 pKa = 11.84DD9 pKa = 3.57ADD11 pKa = 3.74DD12 pKa = 4.2VLFSTSNEE20 pKa = 3.63VTYY23 pKa = 10.59IAFDD27 pKa = 3.82GADD30 pKa = 3.43CKK32 pKa = 10.11YY33 pKa = 9.69TPGDD37 pKa = 3.82CFDD40 pKa = 4.45PAVMTRR46 pKa = 11.84IKK48 pKa = 10.42LSCHH52 pKa = 5.29SRR54 pKa = 11.84LFNATSFDD62 pKa = 4.25QIPEE66 pKa = 4.23LLDD69 pKa = 5.09DD70 pKa = 5.68IIIPDD75 pKa = 3.74YY76 pKa = 11.17CSFGDD81 pKa = 4.56SVDD84 pKa = 3.22INIWIGPSEE93 pKa = 4.39NVSSPYY99 pKa = 10.03FDD101 pKa = 4.0PKK103 pKa = 11.44SNIFAKK109 pKa = 10.65
Molecular weight: 12.18 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A5S6PD00|A0A5S6PD00_BRUMA BMA-MEMB-1 OS=Brugia malayi OX=6279 PE=4 SV=1
MM1 pKa = 7.71LANCYY6 pKa = 7.8WQLDD10 pKa = 3.67HH11 pKa = 6.99QGRR14 pKa = 11.84SRR16 pKa = 11.84KK17 pKa = 9.71EE18 pKa = 3.63GGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGFGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGFGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGFRR1160 pKa = 11.84LRR1162 pKa = 11.84LRR1164 pKa = 11.84LRR1166 pKa = 11.84LRR1168 pKa = 11.84LRR1170 pKa = 11.84LRR1172 pKa = 11.84LRR1174 pKa = 11.84LRR1176 pKa = 11.84LRR1178 pKa = 11.84LRR1180 pKa = 11.84LRR1182 pKa = 11.84LRR1184 pKa = 11.84LRR1186 pKa = 11.84LRR1188 pKa = 11.84LRR1190 pKa = 11.84LRR1192 pKa = 11.84LRR1194 pKa = 11.84LRR1196 pKa = 11.84LRR1198 pKa = 11.84LRR1200 pKa = 11.84LRR1202 pKa = 11.84LRR1204 pKa = 11.84LRR1206 pKa = 11.84LRR1208 pKa = 11.84LRR1210 pKa = 11.84LRR1212 pKa = 11.84LRR1214 pKa = 11.84LRR1216 pKa = 11.84LRR1218 pKa = 11.84LRR1220 pKa = 11.84LRR1222 pKa = 11.84LRR1224 pKa = 11.84LRR1226 pKa = 11.84LRR1228 pKa = 11.84LRR1230 pKa = 11.84LRR1232 pKa = 11.84LRR1234 pKa = 11.84LRR1236 pKa = 11.84LRR1238 pKa = 11.84LRR1240 pKa = 11.84LRR1242 pKa = 11.84LRR1244 pKa = 11.84LRR1246 pKa = 11.84LRR1248 pKa = 11.84LRR1250 pKa = 11.84LRR1252 pKa = 11.84LSLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGFGLGLGLGLGLGLGLGLGLGLGLGLGLGLAA1400 pKa = 4.62
MM1 pKa = 7.71LANCYY6 pKa = 7.8WQLDD10 pKa = 3.67HH11 pKa = 6.99QGRR14 pKa = 11.84SRR16 pKa = 11.84KK17 pKa = 9.71EE18 pKa = 3.63GGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGFGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGFGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGFRR1160 pKa = 11.84LRR1162 pKa = 11.84LRR1164 pKa = 11.84LRR1166 pKa = 11.84LRR1168 pKa = 11.84LRR1170 pKa = 11.84LRR1172 pKa = 11.84LRR1174 pKa = 11.84LRR1176 pKa = 11.84LRR1178 pKa = 11.84LRR1180 pKa = 11.84LRR1182 pKa = 11.84LRR1184 pKa = 11.84LRR1186 pKa = 11.84LRR1188 pKa = 11.84LRR1190 pKa = 11.84LRR1192 pKa = 11.84LRR1194 pKa = 11.84LRR1196 pKa = 11.84LRR1198 pKa = 11.84LRR1200 pKa = 11.84LRR1202 pKa = 11.84LRR1204 pKa = 11.84LRR1206 pKa = 11.84LRR1208 pKa = 11.84LRR1210 pKa = 11.84LRR1212 pKa = 11.84LRR1214 pKa = 11.84LRR1216 pKa = 11.84LRR1218 pKa = 11.84LRR1220 pKa = 11.84LRR1222 pKa = 11.84LRR1224 pKa = 11.84LRR1226 pKa = 11.84LRR1228 pKa = 11.84LRR1230 pKa = 11.84LRR1232 pKa = 11.84LRR1234 pKa = 11.84LRR1236 pKa = 11.84LRR1238 pKa = 11.84LRR1240 pKa = 11.84LRR1242 pKa = 11.84LRR1244 pKa = 11.84LRR1246 pKa = 11.84LRR1248 pKa = 11.84LRR1250 pKa = 11.84LRR1252 pKa = 11.84LSLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGLGFGLGLGLGLGLGLGLGLGLGLGLGLGLGLAA1400 pKa = 4.62
Molecular weight: 124.63 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
4761086 |
15 |
11701 |
466.5 |
52.81 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.109 ± 0.02 | 2.205 ± 0.024 |
5.461 ± 0.018 | 6.685 ± 0.048 |
4.077 ± 0.021 | 5.099 ± 0.037 |
2.426 ± 0.011 | 6.502 ± 0.026 |
6.225 ± 0.035 | 9.235 ± 0.039 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.529 ± 0.013 | 5.167 ± 0.021 |
4.275 ± 0.028 | 4.259 ± 0.025 |
5.673 ± 0.023 | 8.347 ± 0.03 |
5.639 ± 0.02 | 5.863 ± 0.021 |
1.082 ± 0.009 | 3.143 ± 0.016 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
