
Lactobacillus brantae DSM 23927
Taxonomy: cellular organisms; Bacteria; Terrabacteria group; Firmicutes; Bacilli; Lactobacillales; Lactobacillaceae; Lacticaseibacillus; Lacticaseibacillus brantae
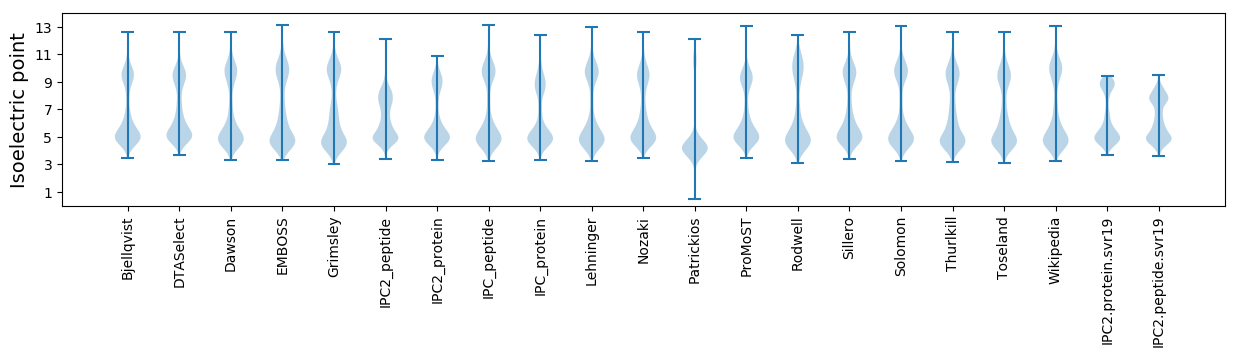
Average proteome isoelectric point is 6.34
Get precalculated fractions of proteins
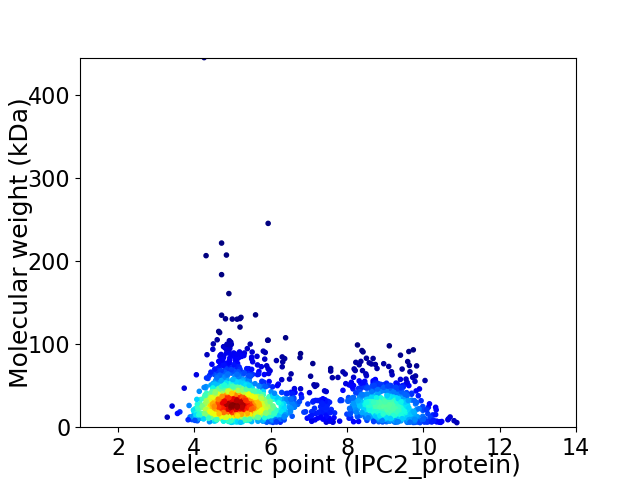
Virtual 2D-PAGE plot for 1863 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0R2BA67|A0A0R2BA67_9LACO M3B subfamily peptidase OS=Lactobacillus brantae DSM 23927 OX=1423727 GN=FC34_GL000783 PE=3 SV=1
MM1 pKa = 7.53SYY3 pKa = 10.56SEE5 pKa = 4.24QMLDD9 pKa = 4.25ALAEE13 pKa = 4.44GNVDD17 pKa = 3.44QANLLFRR24 pKa = 11.84SVLEE28 pKa = 4.5HH29 pKa = 7.77DD30 pKa = 4.96DD31 pKa = 6.15DD32 pKa = 3.88EE33 pKa = 4.84TKK35 pKa = 11.29YY36 pKa = 11.25NLAEE40 pKa = 3.86EE41 pKa = 5.23LYY43 pKa = 10.73ALGFTGQAKK52 pKa = 9.95RR53 pKa = 11.84LYY55 pKa = 8.71QEE57 pKa = 3.94LMGRR61 pKa = 11.84YY62 pKa = 8.68PDD64 pKa = 3.45QDD66 pKa = 4.01DD67 pKa = 3.96IKK69 pKa = 10.39TVLADD74 pKa = 3.36IAVSDD79 pKa = 4.36GDD81 pKa = 3.78PDD83 pKa = 3.56AALNYY88 pKa = 9.87LSKK91 pKa = 10.49IQPDD95 pKa = 3.47SEE97 pKa = 5.32AYY99 pKa = 10.6LEE101 pKa = 4.35SLVTAADD108 pKa = 4.09VYY110 pKa = 9.24QTQGMYY116 pKa = 9.89EE117 pKa = 3.97VSEE120 pKa = 4.22QKK122 pKa = 10.71LLQAQAIAPDD132 pKa = 3.6EE133 pKa = 4.0PVIAFALGEE142 pKa = 4.4LYY144 pKa = 10.38FDD146 pKa = 3.6WGHH149 pKa = 6.0YY150 pKa = 8.96PQAQQAYY157 pKa = 8.91AQLLAAGEE165 pKa = 4.36TEE167 pKa = 4.05LAGVDD172 pKa = 3.6VQARR176 pKa = 11.84LAATLAQLGQYY187 pKa = 9.5EE188 pKa = 4.39DD189 pKa = 5.57AIATYY194 pKa = 9.81EE195 pKa = 3.93ADD197 pKa = 5.5GINQLDD203 pKa = 4.05LNNRR207 pKa = 11.84FQLGGLYY214 pKa = 10.61YY215 pKa = 10.85LVQDD219 pKa = 3.32YY220 pKa = 10.18RR221 pKa = 11.84AAIDD225 pKa = 3.59AFSGVLEE232 pKa = 4.03QDD234 pKa = 2.94QSFASAYY241 pKa = 10.49LPLAQAYY248 pKa = 9.3VADD251 pKa = 4.44NQPDD255 pKa = 3.55MALEE259 pKa = 4.88TIQDD263 pKa = 3.56GVMVDD268 pKa = 3.69DD269 pKa = 4.9TNPDD273 pKa = 3.76LYY275 pKa = 11.53ALGADD280 pKa = 3.96LAIKK284 pKa = 10.62AEE286 pKa = 4.08QNDD289 pKa = 3.64LAEE292 pKa = 4.81NYY294 pKa = 8.99LQKK297 pKa = 11.14ALAIDD302 pKa = 4.09PEE304 pKa = 4.27NLTNALAWSNYY315 pKa = 9.65LYY317 pKa = 10.67LAEE320 pKa = 4.94RR321 pKa = 11.84DD322 pKa = 3.46EE323 pKa = 5.47DD324 pKa = 4.5NIAFLNGLDD333 pKa = 3.87QEE335 pKa = 5.18GEE337 pKa = 4.06VDD339 pKa = 5.01PQINWNLAKK348 pKa = 10.46SEE350 pKa = 3.97ARR352 pKa = 11.84LDD354 pKa = 4.45EE355 pKa = 4.46IDD357 pKa = 3.41QARR360 pKa = 11.84EE361 pKa = 3.7HH362 pKa = 6.56YY363 pKa = 10.75LLAFNRR369 pKa = 11.84LQTQPDD375 pKa = 4.18FLHH378 pKa = 7.3DD379 pKa = 4.69IIDD382 pKa = 4.38FFQMTGAIDD391 pKa = 3.5EE392 pKa = 4.82TKK394 pKa = 10.94AAMQRR399 pKa = 11.84YY400 pKa = 7.95LQLVPDD406 pKa = 4.88DD407 pKa = 5.33LDD409 pKa = 3.37MQEE412 pKa = 4.76RR413 pKa = 11.84YY414 pKa = 10.4DD415 pKa = 4.99RR416 pKa = 11.84LIADD420 pKa = 4.12
MM1 pKa = 7.53SYY3 pKa = 10.56SEE5 pKa = 4.24QMLDD9 pKa = 4.25ALAEE13 pKa = 4.44GNVDD17 pKa = 3.44QANLLFRR24 pKa = 11.84SVLEE28 pKa = 4.5HH29 pKa = 7.77DD30 pKa = 4.96DD31 pKa = 6.15DD32 pKa = 3.88EE33 pKa = 4.84TKK35 pKa = 11.29YY36 pKa = 11.25NLAEE40 pKa = 3.86EE41 pKa = 5.23LYY43 pKa = 10.73ALGFTGQAKK52 pKa = 9.95RR53 pKa = 11.84LYY55 pKa = 8.71QEE57 pKa = 3.94LMGRR61 pKa = 11.84YY62 pKa = 8.68PDD64 pKa = 3.45QDD66 pKa = 4.01DD67 pKa = 3.96IKK69 pKa = 10.39TVLADD74 pKa = 3.36IAVSDD79 pKa = 4.36GDD81 pKa = 3.78PDD83 pKa = 3.56AALNYY88 pKa = 9.87LSKK91 pKa = 10.49IQPDD95 pKa = 3.47SEE97 pKa = 5.32AYY99 pKa = 10.6LEE101 pKa = 4.35SLVTAADD108 pKa = 4.09VYY110 pKa = 9.24QTQGMYY116 pKa = 9.89EE117 pKa = 3.97VSEE120 pKa = 4.22QKK122 pKa = 10.71LLQAQAIAPDD132 pKa = 3.6EE133 pKa = 4.0PVIAFALGEE142 pKa = 4.4LYY144 pKa = 10.38FDD146 pKa = 3.6WGHH149 pKa = 6.0YY150 pKa = 8.96PQAQQAYY157 pKa = 8.91AQLLAAGEE165 pKa = 4.36TEE167 pKa = 4.05LAGVDD172 pKa = 3.6VQARR176 pKa = 11.84LAATLAQLGQYY187 pKa = 9.5EE188 pKa = 4.39DD189 pKa = 5.57AIATYY194 pKa = 9.81EE195 pKa = 3.93ADD197 pKa = 5.5GINQLDD203 pKa = 4.05LNNRR207 pKa = 11.84FQLGGLYY214 pKa = 10.61YY215 pKa = 10.85LVQDD219 pKa = 3.32YY220 pKa = 10.18RR221 pKa = 11.84AAIDD225 pKa = 3.59AFSGVLEE232 pKa = 4.03QDD234 pKa = 2.94QSFASAYY241 pKa = 10.49LPLAQAYY248 pKa = 9.3VADD251 pKa = 4.44NQPDD255 pKa = 3.55MALEE259 pKa = 4.88TIQDD263 pKa = 3.56GVMVDD268 pKa = 3.69DD269 pKa = 4.9TNPDD273 pKa = 3.76LYY275 pKa = 11.53ALGADD280 pKa = 3.96LAIKK284 pKa = 10.62AEE286 pKa = 4.08QNDD289 pKa = 3.64LAEE292 pKa = 4.81NYY294 pKa = 8.99LQKK297 pKa = 11.14ALAIDD302 pKa = 4.09PEE304 pKa = 4.27NLTNALAWSNYY315 pKa = 9.65LYY317 pKa = 10.67LAEE320 pKa = 4.94RR321 pKa = 11.84DD322 pKa = 3.46EE323 pKa = 5.47DD324 pKa = 4.5NIAFLNGLDD333 pKa = 3.87QEE335 pKa = 5.18GEE337 pKa = 4.06VDD339 pKa = 5.01PQINWNLAKK348 pKa = 10.46SEE350 pKa = 3.97ARR352 pKa = 11.84LDD354 pKa = 4.45EE355 pKa = 4.46IDD357 pKa = 3.41QARR360 pKa = 11.84EE361 pKa = 3.7HH362 pKa = 6.56YY363 pKa = 10.75LLAFNRR369 pKa = 11.84LQTQPDD375 pKa = 4.18FLHH378 pKa = 7.3DD379 pKa = 4.69IIDD382 pKa = 4.38FFQMTGAIDD391 pKa = 3.5EE392 pKa = 4.82TKK394 pKa = 10.94AAMQRR399 pKa = 11.84YY400 pKa = 7.95LQLVPDD406 pKa = 4.88DD407 pKa = 5.33LDD409 pKa = 3.37MQEE412 pKa = 4.76RR413 pKa = 11.84YY414 pKa = 10.4DD415 pKa = 4.99RR416 pKa = 11.84LIADD420 pKa = 4.12
Molecular weight: 47.02 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0R2AXI8|A0A0R2AXI8_9LACO 50S ribosomal protein L24 OS=Lactobacillus brantae DSM 23927 OX=1423727 GN=rplX PE=3 SV=1
MM1 pKa = 5.72TTKK4 pKa = 9.85RR5 pKa = 11.84TFQPKK10 pKa = 8.33KK11 pKa = 7.5RR12 pKa = 11.84HH13 pKa = 5.56RR14 pKa = 11.84EE15 pKa = 3.79RR16 pKa = 11.84VHH18 pKa = 7.7GFMKK22 pKa = 10.56RR23 pKa = 11.84MSTKK27 pKa = 10.26NGRR30 pKa = 11.84KK31 pKa = 8.58VLARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 9.94GRR41 pKa = 11.84KK42 pKa = 8.67VLSAA46 pKa = 4.05
MM1 pKa = 5.72TTKK4 pKa = 9.85RR5 pKa = 11.84TFQPKK10 pKa = 8.33KK11 pKa = 7.5RR12 pKa = 11.84HH13 pKa = 5.56RR14 pKa = 11.84EE15 pKa = 3.79RR16 pKa = 11.84VHH18 pKa = 7.7GFMKK22 pKa = 10.56RR23 pKa = 11.84MSTKK27 pKa = 10.26NGRR30 pKa = 11.84KK31 pKa = 8.58VLARR35 pKa = 11.84RR36 pKa = 11.84RR37 pKa = 11.84AKK39 pKa = 9.94GRR41 pKa = 11.84KK42 pKa = 8.67VLSAA46 pKa = 4.05
Molecular weight: 5.52 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
572218 |
46 |
4345 |
307.1 |
33.87 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
9.621 ± 0.06 | 0.214 ± 0.01 |
5.995 ± 0.058 | 4.84 ± 0.059 |
4.204 ± 0.045 | 7.06 ± 0.051 |
2.093 ± 0.031 | 6.443 ± 0.056 |
4.83 ± 0.05 | 10.139 ± 0.078 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
2.634 ± 0.035 | 4.318 ± 0.057 |
4.139 ± 0.035 | 5.118 ± 0.056 |
4.124 ± 0.046 | 5.616 ± 0.056 |
6.665 ± 0.063 | 7.52 ± 0.041 |
1.151 ± 0.025 | 3.273 ± 0.034 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
