
Atkinsonella hypoxylon virus (isolate 2H) (AhV)
Taxonomy: Viruses; Riboviria; Orthornavirae; Pisuviricota; Duplopiviricetes; Durnavirales; Partitiviridae; Betapartitivirus; Atkinsonella hypoxylon virus
Average proteome isoelectric point is 6.4
Get precalculated fractions of proteins
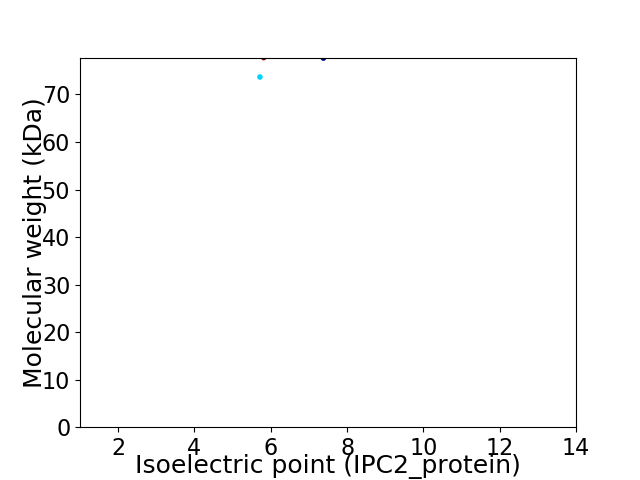
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions


Protein with the lowest isoelectric point:
>sp|Q85056|CAPSD_AHV2H Capsid protein OS=Atkinsonella hypoxylon virus (isolate 2H) OX=647331 PE=3 SV=1
MM1 pKa = 7.61SSNDD5 pKa = 3.3SAQTRR10 pKa = 11.84NLQEE14 pKa = 3.52EE15 pKa = 4.73RR16 pKa = 11.84FNEE19 pKa = 4.26RR20 pKa = 11.84TSTPTVVTAVLPDD33 pKa = 3.8TNGPTTNSTSGSVGPPHH50 pKa = 6.86PTPNVPVPTQSSSDD64 pKa = 3.65PPSASGIFAKK74 pKa = 10.69EE75 pKa = 2.99IDD77 pKa = 3.8LPRR80 pKa = 11.84NVIQHH85 pKa = 6.2SGNKK89 pKa = 9.7FILDD93 pKa = 3.63VVPDD97 pKa = 3.56SRR99 pKa = 11.84FPTFAITEE107 pKa = 4.32FVQRR111 pKa = 11.84SFSNFTFEE119 pKa = 3.95QYY121 pKa = 11.06SYY123 pKa = 11.35VSPASLVGYY132 pKa = 8.97LVYY135 pKa = 10.08MIHH138 pKa = 7.07AFVFLVDD145 pKa = 3.53AFEE148 pKa = 4.54RR149 pKa = 11.84SPMSAYY155 pKa = 10.26ASEE158 pKa = 4.28IDD160 pKa = 3.76ASHH163 pKa = 6.97AYY165 pKa = 10.24LRR167 pKa = 11.84IIDD170 pKa = 4.53AFSDD174 pKa = 3.74AYY176 pKa = 10.1IPDD179 pKa = 3.93FLFEE183 pKa = 4.6ILDD186 pKa = 3.91TYY188 pKa = 11.0LSHH191 pKa = 7.69RR192 pKa = 11.84LDD194 pKa = 3.21IRR196 pKa = 11.84SKK198 pKa = 11.05LEE200 pKa = 3.64MNVSYY205 pKa = 11.17GSVLYY210 pKa = 10.44KK211 pKa = 10.31YY212 pKa = 9.26DD213 pKa = 3.33APRR216 pKa = 11.84IVAPSIFLLAHH227 pKa = 5.99NQLISQSRR235 pKa = 11.84EE236 pKa = 3.35STAYY240 pKa = 9.81EE241 pKa = 3.76KK242 pKa = 10.42WLDD245 pKa = 3.91SIVIHH250 pKa = 6.23YY251 pKa = 9.5SRR253 pKa = 11.84AVIRR257 pKa = 11.84VGNLVGGLYY266 pKa = 10.04QSSHH270 pKa = 6.2GSTTTHH276 pKa = 4.98FTYY279 pKa = 10.71RR280 pKa = 11.84NWFARR285 pKa = 11.84SLSRR289 pKa = 11.84LADD292 pKa = 3.5SATHH296 pKa = 5.66RR297 pKa = 11.84THH299 pKa = 7.14LRR301 pKa = 11.84RR302 pKa = 11.84PMISEE307 pKa = 4.26FDD309 pKa = 3.77YY310 pKa = 11.38NIPSVNNNTYY320 pKa = 10.54NPYY323 pKa = 10.1VHH325 pKa = 7.56LLMLEE330 pKa = 4.09PNNRR334 pKa = 11.84NITLDD339 pKa = 4.09FIRR342 pKa = 11.84SLSSFCSTEE351 pKa = 3.83LKK353 pKa = 9.57ATRR356 pKa = 11.84TLRR359 pKa = 11.84DD360 pKa = 3.65HH361 pKa = 7.12ISRR364 pKa = 11.84RR365 pKa = 11.84SAAISRR371 pKa = 11.84CVIKK375 pKa = 10.88GPEE378 pKa = 3.81APTWHH383 pKa = 6.81SSPLDD388 pKa = 3.72DD389 pKa = 5.66LKK391 pKa = 11.4EE392 pKa = 3.97KK393 pKa = 10.69SKK395 pKa = 10.91QGNFSQFCEE404 pKa = 4.14VAKK407 pKa = 10.73FGLPRR412 pKa = 11.84KK413 pKa = 9.11EE414 pKa = 3.94NSEE417 pKa = 4.18SYY419 pKa = 9.74TFKK422 pKa = 10.65FPKK425 pKa = 9.96DD426 pKa = 3.06ASTIDD431 pKa = 3.17TAFYY435 pKa = 10.31LIQEE439 pKa = 4.42NGRR442 pKa = 11.84SSVLDD447 pKa = 3.28PTTADD452 pKa = 3.56EE453 pKa = 4.31EE454 pKa = 4.62LHH456 pKa = 5.62TEE458 pKa = 4.27GMNLLFDD465 pKa = 5.17PYY467 pKa = 10.98DD468 pKa = 4.44DD469 pKa = 4.51EE470 pKa = 6.63SSAHH474 pKa = 5.68YY475 pKa = 8.7ATVLSGKK482 pKa = 10.14LIQNSNIDD490 pKa = 3.89GEE492 pKa = 4.8TLLLPDD498 pKa = 4.47PTTGLARR505 pKa = 11.84TNSRR509 pKa = 11.84YY510 pKa = 9.78LQGSVLIRR518 pKa = 11.84NVLPEE523 pKa = 4.17FDD525 pKa = 3.2QHH527 pKa = 6.77EE528 pKa = 4.27IRR530 pKa = 11.84LFPRR534 pKa = 11.84YY535 pKa = 8.53PQISRR540 pKa = 11.84LSASLTLLFNMRR552 pKa = 11.84QVWIPRR558 pKa = 11.84FKK560 pKa = 10.91QKK562 pKa = 10.37VDD564 pKa = 3.66EE565 pKa = 4.23QPKK568 pKa = 10.35LSNFSWNEE576 pKa = 3.81GCDD579 pKa = 3.51GTVPSLNVVTAEE591 pKa = 4.14SSTNGPAAEE600 pKa = 3.97QQVILWSSYY609 pKa = 7.78RR610 pKa = 11.84HH611 pKa = 5.58VSNSDD616 pKa = 3.0RR617 pKa = 11.84PTVDD621 pKa = 2.39TVYY624 pKa = 10.86YY625 pKa = 10.37YY626 pKa = 10.07STLEE630 pKa = 3.97LLFGTRR636 pKa = 11.84SSMMQTYY643 pKa = 10.32NLHH646 pKa = 5.83QLLSLHH652 pKa = 6.42
MM1 pKa = 7.61SSNDD5 pKa = 3.3SAQTRR10 pKa = 11.84NLQEE14 pKa = 3.52EE15 pKa = 4.73RR16 pKa = 11.84FNEE19 pKa = 4.26RR20 pKa = 11.84TSTPTVVTAVLPDD33 pKa = 3.8TNGPTTNSTSGSVGPPHH50 pKa = 6.86PTPNVPVPTQSSSDD64 pKa = 3.65PPSASGIFAKK74 pKa = 10.69EE75 pKa = 2.99IDD77 pKa = 3.8LPRR80 pKa = 11.84NVIQHH85 pKa = 6.2SGNKK89 pKa = 9.7FILDD93 pKa = 3.63VVPDD97 pKa = 3.56SRR99 pKa = 11.84FPTFAITEE107 pKa = 4.32FVQRR111 pKa = 11.84SFSNFTFEE119 pKa = 3.95QYY121 pKa = 11.06SYY123 pKa = 11.35VSPASLVGYY132 pKa = 8.97LVYY135 pKa = 10.08MIHH138 pKa = 7.07AFVFLVDD145 pKa = 3.53AFEE148 pKa = 4.54RR149 pKa = 11.84SPMSAYY155 pKa = 10.26ASEE158 pKa = 4.28IDD160 pKa = 3.76ASHH163 pKa = 6.97AYY165 pKa = 10.24LRR167 pKa = 11.84IIDD170 pKa = 4.53AFSDD174 pKa = 3.74AYY176 pKa = 10.1IPDD179 pKa = 3.93FLFEE183 pKa = 4.6ILDD186 pKa = 3.91TYY188 pKa = 11.0LSHH191 pKa = 7.69RR192 pKa = 11.84LDD194 pKa = 3.21IRR196 pKa = 11.84SKK198 pKa = 11.05LEE200 pKa = 3.64MNVSYY205 pKa = 11.17GSVLYY210 pKa = 10.44KK211 pKa = 10.31YY212 pKa = 9.26DD213 pKa = 3.33APRR216 pKa = 11.84IVAPSIFLLAHH227 pKa = 5.99NQLISQSRR235 pKa = 11.84EE236 pKa = 3.35STAYY240 pKa = 9.81EE241 pKa = 3.76KK242 pKa = 10.42WLDD245 pKa = 3.91SIVIHH250 pKa = 6.23YY251 pKa = 9.5SRR253 pKa = 11.84AVIRR257 pKa = 11.84VGNLVGGLYY266 pKa = 10.04QSSHH270 pKa = 6.2GSTTTHH276 pKa = 4.98FTYY279 pKa = 10.71RR280 pKa = 11.84NWFARR285 pKa = 11.84SLSRR289 pKa = 11.84LADD292 pKa = 3.5SATHH296 pKa = 5.66RR297 pKa = 11.84THH299 pKa = 7.14LRR301 pKa = 11.84RR302 pKa = 11.84PMISEE307 pKa = 4.26FDD309 pKa = 3.77YY310 pKa = 11.38NIPSVNNNTYY320 pKa = 10.54NPYY323 pKa = 10.1VHH325 pKa = 7.56LLMLEE330 pKa = 4.09PNNRR334 pKa = 11.84NITLDD339 pKa = 4.09FIRR342 pKa = 11.84SLSSFCSTEE351 pKa = 3.83LKK353 pKa = 9.57ATRR356 pKa = 11.84TLRR359 pKa = 11.84DD360 pKa = 3.65HH361 pKa = 7.12ISRR364 pKa = 11.84RR365 pKa = 11.84SAAISRR371 pKa = 11.84CVIKK375 pKa = 10.88GPEE378 pKa = 3.81APTWHH383 pKa = 6.81SSPLDD388 pKa = 3.72DD389 pKa = 5.66LKK391 pKa = 11.4EE392 pKa = 3.97KK393 pKa = 10.69SKK395 pKa = 10.91QGNFSQFCEE404 pKa = 4.14VAKK407 pKa = 10.73FGLPRR412 pKa = 11.84KK413 pKa = 9.11EE414 pKa = 3.94NSEE417 pKa = 4.18SYY419 pKa = 9.74TFKK422 pKa = 10.65FPKK425 pKa = 9.96DD426 pKa = 3.06ASTIDD431 pKa = 3.17TAFYY435 pKa = 10.31LIQEE439 pKa = 4.42NGRR442 pKa = 11.84SSVLDD447 pKa = 3.28PTTADD452 pKa = 3.56EE453 pKa = 4.31EE454 pKa = 4.62LHH456 pKa = 5.62TEE458 pKa = 4.27GMNLLFDD465 pKa = 5.17PYY467 pKa = 10.98DD468 pKa = 4.44DD469 pKa = 4.51EE470 pKa = 6.63SSAHH474 pKa = 5.68YY475 pKa = 8.7ATVLSGKK482 pKa = 10.14LIQNSNIDD490 pKa = 3.89GEE492 pKa = 4.8TLLLPDD498 pKa = 4.47PTTGLARR505 pKa = 11.84TNSRR509 pKa = 11.84YY510 pKa = 9.78LQGSVLIRR518 pKa = 11.84NVLPEE523 pKa = 4.17FDD525 pKa = 3.2QHH527 pKa = 6.77EE528 pKa = 4.27IRR530 pKa = 11.84LFPRR534 pKa = 11.84YY535 pKa = 8.53PQISRR540 pKa = 11.84LSASLTLLFNMRR552 pKa = 11.84QVWIPRR558 pKa = 11.84FKK560 pKa = 10.91QKK562 pKa = 10.37VDD564 pKa = 3.66EE565 pKa = 4.23QPKK568 pKa = 10.35LSNFSWNEE576 pKa = 3.81GCDD579 pKa = 3.51GTVPSLNVVTAEE591 pKa = 4.14SSTNGPAAEE600 pKa = 3.97QQVILWSSYY609 pKa = 7.78RR610 pKa = 11.84HH611 pKa = 5.58VSNSDD616 pKa = 3.0RR617 pKa = 11.84PTVDD621 pKa = 2.39TVYY624 pKa = 10.86YY625 pKa = 10.37YY626 pKa = 10.07STLEE630 pKa = 3.97LLFGTRR636 pKa = 11.84SSMMQTYY643 pKa = 10.32NLHH646 pKa = 5.83QLLSLHH652 pKa = 6.42
Molecular weight: 73.67 kDa
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>sp|Q85056|CAPSD_AHV2H Capsid protein OS=Atkinsonella hypoxylon virus (isolate 2H) OX=647331 PE=3 SV=1
MM1 pKa = 7.07STLLIPQDD9 pKa = 4.27TIAHH13 pKa = 5.87TFDD16 pKa = 3.27EE17 pKa = 4.97AVASEE22 pKa = 4.4SNLRR26 pKa = 11.84IDD28 pKa = 4.26EE29 pKa = 4.2VPEE32 pKa = 4.03NYY34 pKa = 10.08LEE36 pKa = 4.59RR37 pKa = 11.84FIHH40 pKa = 6.43PSEE43 pKa = 4.38PEE45 pKa = 3.62NFEE48 pKa = 4.29FYY50 pKa = 10.77SLRR53 pKa = 11.84DD54 pKa = 2.96SDD56 pKa = 4.19IPSKK60 pKa = 10.73RR61 pKa = 11.84IPKK64 pKa = 10.14NGIQVFEE71 pKa = 4.01NLKK74 pKa = 9.96YY75 pKa = 9.46HH76 pKa = 6.24TNSKK80 pKa = 11.06DD81 pKa = 3.56NLYY84 pKa = 10.65KK85 pKa = 10.52DD86 pKa = 3.98QPSSGPSPMRR96 pKa = 11.84GVANIIRR103 pKa = 11.84EE104 pKa = 4.22YY105 pKa = 10.42FPQYY109 pKa = 11.01LDD111 pKa = 6.1DD112 pKa = 4.31LRR114 pKa = 11.84TWCRR118 pKa = 11.84PKK120 pKa = 10.95SSDD123 pKa = 4.36DD124 pKa = 4.37SIFNDD129 pKa = 3.73FNHH132 pKa = 6.22EE133 pKa = 3.93QRR135 pKa = 11.84ITQPFTEE142 pKa = 4.15EE143 pKa = 3.77RR144 pKa = 11.84EE145 pKa = 4.09RR146 pKa = 11.84RR147 pKa = 11.84LLPLIDD153 pKa = 4.15HH154 pKa = 7.03FLGIKK159 pKa = 10.0PYY161 pKa = 10.62DD162 pKa = 3.38IVHH165 pKa = 5.78YY166 pKa = 9.74CDD168 pKa = 2.86TRR170 pKa = 11.84FYY172 pKa = 10.09PWKK175 pKa = 10.03LSTRR179 pKa = 11.84ADD181 pKa = 3.59YY182 pKa = 10.26FHH184 pKa = 6.62NHH186 pKa = 4.37SRR188 pKa = 11.84DD189 pKa = 3.68RR190 pKa = 11.84KK191 pKa = 8.09AHH193 pKa = 5.81AAKK196 pKa = 10.14SHH198 pKa = 7.02PDD200 pKa = 3.31FATGPTKK207 pKa = 10.32KK208 pKa = 10.42SYY210 pKa = 10.66FINSHH215 pKa = 6.13LFFDD219 pKa = 4.61RR220 pKa = 11.84STVHH224 pKa = 6.64NIKK227 pKa = 10.5EE228 pKa = 3.97YY229 pKa = 10.7GFPFRR234 pKa = 11.84PTTDD238 pKa = 2.82SARR241 pKa = 11.84NEE243 pKa = 4.18TLLDD247 pKa = 3.36LWFKK251 pKa = 10.71KK252 pKa = 10.78VPTEE256 pKa = 3.72LLVRR260 pKa = 11.84SHH262 pKa = 6.88ISKK265 pKa = 10.4RR266 pKa = 11.84DD267 pKa = 3.6NLKK270 pKa = 10.07VRR272 pKa = 11.84PVYY275 pKa = 9.36NAPMIYY281 pKa = 9.79IRR283 pKa = 11.84IEE285 pKa = 3.88CMLFYY290 pKa = 10.57PLLAQARR297 pKa = 11.84KK298 pKa = 9.35RR299 pKa = 11.84DD300 pKa = 3.66CCIMYY305 pKa = 10.45GLEE308 pKa = 4.21TIRR311 pKa = 11.84GGMNEE316 pKa = 4.62LEE318 pKa = 4.6RR319 pKa = 11.84ISNAFNSFLLIDD331 pKa = 4.18WSRR334 pKa = 11.84FDD336 pKa = 3.94HH337 pKa = 6.72LAPFTISNFFFKK349 pKa = 10.19KK350 pKa = 9.4WLPTKK355 pKa = 10.38ILIDD359 pKa = 3.39HH360 pKa = 7.16GYY362 pKa = 9.68AQISNYY368 pKa = 8.82HH369 pKa = 5.92DD370 pKa = 3.79HH371 pKa = 5.95VHH373 pKa = 6.1SFSAQAQSHH382 pKa = 7.03GIPMISKK389 pKa = 9.5EE390 pKa = 4.14YY391 pKa = 8.18QTPPEE396 pKa = 3.92ATVFAKK402 pKa = 10.34KK403 pKa = 10.25VLNLISFLEE412 pKa = 3.74RR413 pKa = 11.84WYY415 pKa = 10.94RR416 pKa = 11.84DD417 pKa = 3.53MVFVTPDD424 pKa = 2.73GFAYY428 pKa = 10.29RR429 pKa = 11.84RR430 pKa = 11.84THH432 pKa = 6.41AGVPSGILMTQFIDD446 pKa = 3.59SFVNLTILLDD456 pKa = 3.63GLIEE460 pKa = 4.64FGFTDD465 pKa = 4.07EE466 pKa = 5.17EE467 pKa = 4.4IKK469 pKa = 10.79QLLVFIMGDD478 pKa = 3.48DD479 pKa = 3.72NVIFTPWTLLKK490 pKa = 10.74LIEE493 pKa = 4.47FFDD496 pKa = 3.67WFAKK500 pKa = 8.65YY501 pKa = 8.85TLDD504 pKa = 4.06RR505 pKa = 11.84FGMVINISKK514 pKa = 10.55SAVTSIRR521 pKa = 11.84RR522 pKa = 11.84KK523 pKa = 9.97IEE525 pKa = 3.55VLGYY529 pKa = 7.72TNNYY533 pKa = 8.69GFPTRR538 pKa = 11.84SISKK542 pKa = 10.27LVGQLAYY549 pKa = 9.68PEE551 pKa = 4.23RR552 pKa = 11.84HH553 pKa = 5.03VTDD556 pKa = 4.87ADD558 pKa = 3.16MCMRR562 pKa = 11.84AIGFAYY568 pKa = 10.01ASCAQSEE575 pKa = 4.66TFHH578 pKa = 6.83ALCKK582 pKa = 10.35KK583 pKa = 9.16VFQYY587 pKa = 11.09YY588 pKa = 9.35FAKK591 pKa = 10.12TSINEE596 pKa = 3.85RR597 pKa = 11.84LILKK601 pKa = 9.13GRR603 pKa = 11.84KK604 pKa = 9.12AEE606 pKa = 3.95LPGMFFAYY614 pKa = 9.6PDD616 pKa = 3.53VSEE619 pKa = 5.43HH620 pKa = 5.91IRR622 pKa = 11.84LDD624 pKa = 3.54HH625 pKa = 6.68FPSLSEE631 pKa = 4.1VRR633 pKa = 11.84ILLSKK638 pKa = 9.78FQGYY642 pKa = 9.62LKK644 pKa = 7.84EE645 pKa = 4.59TPFGTIPTFSTPQTLRR661 pKa = 11.84DD662 pKa = 3.51QTQQ665 pKa = 2.43
MM1 pKa = 7.07STLLIPQDD9 pKa = 4.27TIAHH13 pKa = 5.87TFDD16 pKa = 3.27EE17 pKa = 4.97AVASEE22 pKa = 4.4SNLRR26 pKa = 11.84IDD28 pKa = 4.26EE29 pKa = 4.2VPEE32 pKa = 4.03NYY34 pKa = 10.08LEE36 pKa = 4.59RR37 pKa = 11.84FIHH40 pKa = 6.43PSEE43 pKa = 4.38PEE45 pKa = 3.62NFEE48 pKa = 4.29FYY50 pKa = 10.77SLRR53 pKa = 11.84DD54 pKa = 2.96SDD56 pKa = 4.19IPSKK60 pKa = 10.73RR61 pKa = 11.84IPKK64 pKa = 10.14NGIQVFEE71 pKa = 4.01NLKK74 pKa = 9.96YY75 pKa = 9.46HH76 pKa = 6.24TNSKK80 pKa = 11.06DD81 pKa = 3.56NLYY84 pKa = 10.65KK85 pKa = 10.52DD86 pKa = 3.98QPSSGPSPMRR96 pKa = 11.84GVANIIRR103 pKa = 11.84EE104 pKa = 4.22YY105 pKa = 10.42FPQYY109 pKa = 11.01LDD111 pKa = 6.1DD112 pKa = 4.31LRR114 pKa = 11.84TWCRR118 pKa = 11.84PKK120 pKa = 10.95SSDD123 pKa = 4.36DD124 pKa = 4.37SIFNDD129 pKa = 3.73FNHH132 pKa = 6.22EE133 pKa = 3.93QRR135 pKa = 11.84ITQPFTEE142 pKa = 4.15EE143 pKa = 3.77RR144 pKa = 11.84EE145 pKa = 4.09RR146 pKa = 11.84RR147 pKa = 11.84LLPLIDD153 pKa = 4.15HH154 pKa = 7.03FLGIKK159 pKa = 10.0PYY161 pKa = 10.62DD162 pKa = 3.38IVHH165 pKa = 5.78YY166 pKa = 9.74CDD168 pKa = 2.86TRR170 pKa = 11.84FYY172 pKa = 10.09PWKK175 pKa = 10.03LSTRR179 pKa = 11.84ADD181 pKa = 3.59YY182 pKa = 10.26FHH184 pKa = 6.62NHH186 pKa = 4.37SRR188 pKa = 11.84DD189 pKa = 3.68RR190 pKa = 11.84KK191 pKa = 8.09AHH193 pKa = 5.81AAKK196 pKa = 10.14SHH198 pKa = 7.02PDD200 pKa = 3.31FATGPTKK207 pKa = 10.32KK208 pKa = 10.42SYY210 pKa = 10.66FINSHH215 pKa = 6.13LFFDD219 pKa = 4.61RR220 pKa = 11.84STVHH224 pKa = 6.64NIKK227 pKa = 10.5EE228 pKa = 3.97YY229 pKa = 10.7GFPFRR234 pKa = 11.84PTTDD238 pKa = 2.82SARR241 pKa = 11.84NEE243 pKa = 4.18TLLDD247 pKa = 3.36LWFKK251 pKa = 10.71KK252 pKa = 10.78VPTEE256 pKa = 3.72LLVRR260 pKa = 11.84SHH262 pKa = 6.88ISKK265 pKa = 10.4RR266 pKa = 11.84DD267 pKa = 3.6NLKK270 pKa = 10.07VRR272 pKa = 11.84PVYY275 pKa = 9.36NAPMIYY281 pKa = 9.79IRR283 pKa = 11.84IEE285 pKa = 3.88CMLFYY290 pKa = 10.57PLLAQARR297 pKa = 11.84KK298 pKa = 9.35RR299 pKa = 11.84DD300 pKa = 3.66CCIMYY305 pKa = 10.45GLEE308 pKa = 4.21TIRR311 pKa = 11.84GGMNEE316 pKa = 4.62LEE318 pKa = 4.6RR319 pKa = 11.84ISNAFNSFLLIDD331 pKa = 4.18WSRR334 pKa = 11.84FDD336 pKa = 3.94HH337 pKa = 6.72LAPFTISNFFFKK349 pKa = 10.19KK350 pKa = 9.4WLPTKK355 pKa = 10.38ILIDD359 pKa = 3.39HH360 pKa = 7.16GYY362 pKa = 9.68AQISNYY368 pKa = 8.82HH369 pKa = 5.92DD370 pKa = 3.79HH371 pKa = 5.95VHH373 pKa = 6.1SFSAQAQSHH382 pKa = 7.03GIPMISKK389 pKa = 9.5EE390 pKa = 4.14YY391 pKa = 8.18QTPPEE396 pKa = 3.92ATVFAKK402 pKa = 10.34KK403 pKa = 10.25VLNLISFLEE412 pKa = 3.74RR413 pKa = 11.84WYY415 pKa = 10.94RR416 pKa = 11.84DD417 pKa = 3.53MVFVTPDD424 pKa = 2.73GFAYY428 pKa = 10.29RR429 pKa = 11.84RR430 pKa = 11.84THH432 pKa = 6.41AGVPSGILMTQFIDD446 pKa = 3.59SFVNLTILLDD456 pKa = 3.63GLIEE460 pKa = 4.64FGFTDD465 pKa = 4.07EE466 pKa = 5.17EE467 pKa = 4.4IKK469 pKa = 10.79QLLVFIMGDD478 pKa = 3.48DD479 pKa = 3.72NVIFTPWTLLKK490 pKa = 10.74LIEE493 pKa = 4.47FFDD496 pKa = 3.67WFAKK500 pKa = 8.65YY501 pKa = 8.85TLDD504 pKa = 4.06RR505 pKa = 11.84FGMVINISKK514 pKa = 10.55SAVTSIRR521 pKa = 11.84RR522 pKa = 11.84KK523 pKa = 9.97IEE525 pKa = 3.55VLGYY529 pKa = 7.72TNNYY533 pKa = 8.69GFPTRR538 pKa = 11.84SISKK542 pKa = 10.27LVGQLAYY549 pKa = 9.68PEE551 pKa = 4.23RR552 pKa = 11.84HH553 pKa = 5.03VTDD556 pKa = 4.87ADD558 pKa = 3.16MCMRR562 pKa = 11.84AIGFAYY568 pKa = 10.01ASCAQSEE575 pKa = 4.66TFHH578 pKa = 6.83ALCKK582 pKa = 10.35KK583 pKa = 9.16VFQYY587 pKa = 11.09YY588 pKa = 9.35FAKK591 pKa = 10.12TSINEE596 pKa = 3.85RR597 pKa = 11.84LILKK601 pKa = 9.13GRR603 pKa = 11.84KK604 pKa = 9.12AEE606 pKa = 3.95LPGMFFAYY614 pKa = 9.6PDD616 pKa = 3.53VSEE619 pKa = 5.43HH620 pKa = 5.91IRR622 pKa = 11.84LDD624 pKa = 3.54HH625 pKa = 6.68FPSLSEE631 pKa = 4.1VRR633 pKa = 11.84ILLSKK638 pKa = 9.78FQGYY642 pKa = 9.62LKK644 pKa = 7.84EE645 pKa = 4.59TPFGTIPTFSTPQTLRR661 pKa = 11.84DD662 pKa = 3.51QTQQ665 pKa = 2.43
Molecular weight: 77.65 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
1317 |
652 |
665 |
658.5 |
75.66 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
5.391 ± 0.186 | 0.911 ± 0.195 |
5.771 ± 0.264 | 5.239 ± 0.117 |
6.606 ± 0.913 | 3.797 ± 0.076 |
3.341 ± 0.179 | 6.378 ± 0.763 |
4.176 ± 0.929 | 9.112 ± 0.261 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
1.822 ± 0.189 | 4.86 ± 0.434 |
5.998 ± 0.09 | 3.265 ± 0.172 |
6.302 ± 0.11 | 9.719 ± 1.674 |
6.758 ± 0.396 | 5.011 ± 0.637 |
1.063 ± 0.094 | 4.48 ± 0.021 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
