
Avon-Heathcote Estuary associated circular virus 6
Taxonomy: Viruses; Monodnaviria; Shotokuvirae; Cressdnaviricota; Arfiviricetes; Cirlivirales; Circoviridae; unclassified Circoviridae
Average proteome isoelectric point is 8.39
Get precalculated fractions of proteins
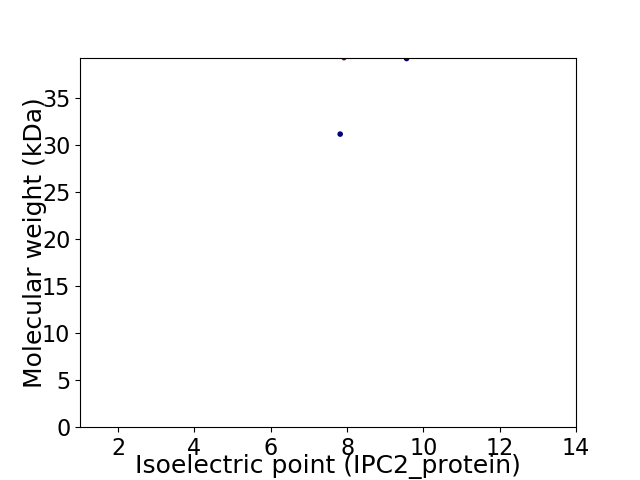
Virtual 2D-PAGE plot for 2 proteins (isoelectric point calculated using IPC2_protein)
Get csv file with sequences according to given criteria:
* You can choose from 21 different methods for calculating isoelectric point
Summary statistics related to proteome-wise predictions
Protein with the lowest isoelectric point:
>tr|A0A0C5IBI9|A0A0C5IBI9_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 6 OX=1618257 PE=3 SV=1
MM1 pKa = 7.17SRR3 pKa = 11.84SRR5 pKa = 11.84NWCFTLHH12 pKa = 6.85NYY14 pKa = 9.15TDD16 pKa = 4.09DD17 pKa = 5.34DD18 pKa = 4.0IEE20 pKa = 3.91IFKK23 pKa = 10.6NIKK26 pKa = 9.07CRR28 pKa = 11.84YY29 pKa = 7.63IIYY32 pKa = 10.09GKK34 pKa = 9.18EE35 pKa = 3.33ICPTTKK41 pKa = 9.88RR42 pKa = 11.84QHH44 pKa = 5.2LQGYY48 pKa = 8.21IQFDD52 pKa = 3.59NQRR55 pKa = 11.84TITAVKK61 pKa = 10.13KK62 pKa = 10.5FLGLPKK68 pKa = 9.95IHH70 pKa = 7.29LEE72 pKa = 3.92IANGSSDD79 pKa = 4.57DD80 pKa = 3.79NKK82 pKa = 10.59TYY84 pKa = 10.49CSKK87 pKa = 11.13DD88 pKa = 2.49GDD90 pKa = 3.73MYY92 pKa = 11.43EE93 pKa = 5.73RR94 pKa = 11.84GDD96 pKa = 3.75CKK98 pKa = 10.5QQGRR102 pKa = 11.84RR103 pKa = 11.84TDD105 pKa = 2.95ITDD108 pKa = 3.29IKK110 pKa = 11.65DD111 pKa = 3.61MISNGATMRR120 pKa = 11.84DD121 pKa = 4.1IIPHH125 pKa = 5.29ATSVQSVRR133 pKa = 11.84MAEE136 pKa = 3.62IHH138 pKa = 6.56LKK140 pKa = 10.5YY141 pKa = 10.43SEE143 pKa = 4.7QKK145 pKa = 9.32RR146 pKa = 11.84TWKK149 pKa = 10.4PKK151 pKa = 8.07VQWFYY156 pKa = 11.71GPTGTGKK163 pKa = 8.11TRR165 pKa = 11.84TAYY168 pKa = 9.86EE169 pKa = 4.11ILEE172 pKa = 4.46DD173 pKa = 4.79PYY175 pKa = 9.42TTLDD179 pKa = 3.12TGQWWEE185 pKa = 4.65GYY187 pKa = 8.95DD188 pKa = 3.2AHH190 pKa = 6.79EE191 pKa = 4.34NVIIDD196 pKa = 4.0DD197 pKa = 3.74MRR199 pKa = 11.84GDD201 pKa = 3.53FMKK204 pKa = 10.99YY205 pKa = 9.71HH206 pKa = 6.27VLLKK210 pKa = 10.97LLDD213 pKa = 3.82RR214 pKa = 11.84YY215 pKa = 10.95AYY217 pKa = 9.52IVEE220 pKa = 4.4CKK222 pKa = 10.35GGSRR226 pKa = 11.84QFLARR231 pKa = 11.84HH232 pKa = 6.25IIITSAFHH240 pKa = 6.54PKK242 pKa = 10.36DD243 pKa = 3.55VFHH246 pKa = 6.44TRR248 pKa = 11.84EE249 pKa = 4.8DD250 pKa = 3.19IAQLMRR256 pKa = 11.84RR257 pKa = 11.84IDD259 pKa = 3.59CVKK262 pKa = 10.24EE263 pKa = 4.68FKK265 pKa = 10.82
MM1 pKa = 7.17SRR3 pKa = 11.84SRR5 pKa = 11.84NWCFTLHH12 pKa = 6.85NYY14 pKa = 9.15TDD16 pKa = 4.09DD17 pKa = 5.34DD18 pKa = 4.0IEE20 pKa = 3.91IFKK23 pKa = 10.6NIKK26 pKa = 9.07CRR28 pKa = 11.84YY29 pKa = 7.63IIYY32 pKa = 10.09GKK34 pKa = 9.18EE35 pKa = 3.33ICPTTKK41 pKa = 9.88RR42 pKa = 11.84QHH44 pKa = 5.2LQGYY48 pKa = 8.21IQFDD52 pKa = 3.59NQRR55 pKa = 11.84TITAVKK61 pKa = 10.13KK62 pKa = 10.5FLGLPKK68 pKa = 9.95IHH70 pKa = 7.29LEE72 pKa = 3.92IANGSSDD79 pKa = 4.57DD80 pKa = 3.79NKK82 pKa = 10.59TYY84 pKa = 10.49CSKK87 pKa = 11.13DD88 pKa = 2.49GDD90 pKa = 3.73MYY92 pKa = 11.43EE93 pKa = 5.73RR94 pKa = 11.84GDD96 pKa = 3.75CKK98 pKa = 10.5QQGRR102 pKa = 11.84RR103 pKa = 11.84TDD105 pKa = 2.95ITDD108 pKa = 3.29IKK110 pKa = 11.65DD111 pKa = 3.61MISNGATMRR120 pKa = 11.84DD121 pKa = 4.1IIPHH125 pKa = 5.29ATSVQSVRR133 pKa = 11.84MAEE136 pKa = 3.62IHH138 pKa = 6.56LKK140 pKa = 10.5YY141 pKa = 10.43SEE143 pKa = 4.7QKK145 pKa = 9.32RR146 pKa = 11.84TWKK149 pKa = 10.4PKK151 pKa = 8.07VQWFYY156 pKa = 11.71GPTGTGKK163 pKa = 8.11TRR165 pKa = 11.84TAYY168 pKa = 9.86EE169 pKa = 4.11ILEE172 pKa = 4.46DD173 pKa = 4.79PYY175 pKa = 9.42TTLDD179 pKa = 3.12TGQWWEE185 pKa = 4.65GYY187 pKa = 8.95DD188 pKa = 3.2AHH190 pKa = 6.79EE191 pKa = 4.34NVIIDD196 pKa = 4.0DD197 pKa = 3.74MRR199 pKa = 11.84GDD201 pKa = 3.53FMKK204 pKa = 10.99YY205 pKa = 9.71HH206 pKa = 6.27VLLKK210 pKa = 10.97LLDD213 pKa = 3.82RR214 pKa = 11.84YY215 pKa = 10.95AYY217 pKa = 9.52IVEE220 pKa = 4.4CKK222 pKa = 10.35GGSRR226 pKa = 11.84QFLARR231 pKa = 11.84HH232 pKa = 6.25IIITSAFHH240 pKa = 6.54PKK242 pKa = 10.36DD243 pKa = 3.55VFHH246 pKa = 6.44TRR248 pKa = 11.84EE249 pKa = 4.8DD250 pKa = 3.19IAQLMRR256 pKa = 11.84RR257 pKa = 11.84IDD259 pKa = 3.59CVKK262 pKa = 10.24EE263 pKa = 4.68FKK265 pKa = 10.82
Molecular weight: 31.17 kDa
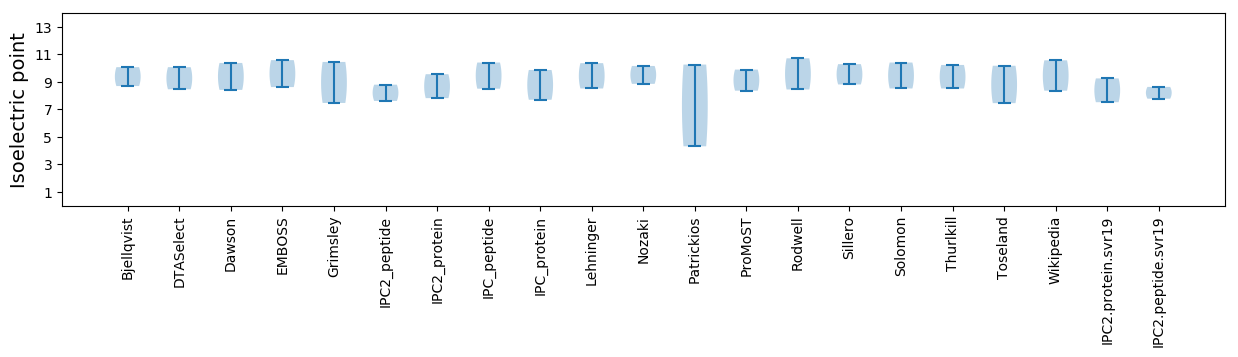
Isoelectric point according different methods:
Protein with the highest isoelectric point:
>tr|A0A0C5IBI9|A0A0C5IBI9_9CIRC ATP-dependent helicase Rep OS=Avon-Heathcote Estuary associated circular virus 6 OX=1618257 PE=3 SV=1
MM1 pKa = 7.52PPIRR5 pKa = 11.84KK6 pKa = 9.29LDD8 pKa = 3.66TRR10 pKa = 11.84AKK12 pKa = 9.72RR13 pKa = 11.84AAALRR18 pKa = 11.84RR19 pKa = 11.84GQIRR23 pKa = 11.84APRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84GAPFRR33 pKa = 11.84YY34 pKa = 8.55RR35 pKa = 11.84TGTKK39 pKa = 9.86RR40 pKa = 11.84GRR42 pKa = 11.84TTAARR47 pKa = 11.84RR48 pKa = 11.84NGNTIALTTLSRR60 pKa = 11.84KK61 pKa = 8.65QEE63 pKa = 4.04NLSISYY69 pKa = 10.75RR70 pKa = 11.84EE71 pKa = 3.73MLEE74 pKa = 4.01FNDD77 pKa = 3.69MGGDD81 pKa = 3.61NGSTPCLIRR90 pKa = 11.84VNLNNPVIGGATPTGPDD107 pKa = 3.39TIVSVVGNLKK117 pKa = 9.46TGATDD122 pKa = 3.33PTFIPHH128 pKa = 6.36SYY130 pKa = 9.33EE131 pKa = 3.8NKK133 pKa = 9.95RR134 pKa = 11.84NLSDD138 pKa = 3.66RR139 pKa = 11.84LAEE142 pKa = 3.95YY143 pKa = 10.37FSIYY147 pKa = 8.66RR148 pKa = 11.84TAIVTSAEE156 pKa = 4.19VTVVCTPKK164 pKa = 10.62LNQLNGMTGNNRR176 pKa = 11.84SIVPFTQNRR185 pKa = 11.84ASEE188 pKa = 4.36DD189 pKa = 3.53PAVAGQATYY198 pKa = 10.5LYY200 pKa = 9.73QHH202 pKa = 6.89NANAMPQVEE211 pKa = 4.64VWSIRR216 pKa = 11.84QQNQGQLTSAAAGSPPLEE234 pKa = 4.22TLKK237 pKa = 10.84QGIPGMRR244 pKa = 11.84MTRR247 pKa = 11.84LNVVPNSTKK256 pKa = 10.13GVTYY260 pKa = 10.74KK261 pKa = 10.09MSYY264 pKa = 8.16TPKK267 pKa = 10.56SMYY270 pKa = 10.55AISDD274 pKa = 3.21WKK276 pKa = 11.0DD277 pKa = 3.04NKK279 pKa = 10.45KK280 pKa = 10.0VLKK283 pKa = 10.3IFNNAVKK290 pKa = 10.84NPDD293 pKa = 3.11QKK295 pKa = 10.48EE296 pKa = 3.66AYY298 pKa = 8.76MYY300 pKa = 10.82VGIAGRR306 pKa = 11.84FQGLDD311 pKa = 3.45PVASLANMGLPHH323 pKa = 6.72FNVEE327 pKa = 4.19VKK329 pKa = 10.54VKK331 pKa = 10.45YY332 pKa = 10.38NINFSEE338 pKa = 4.94RR339 pKa = 11.84YY340 pKa = 10.02NIDD343 pKa = 3.3GNNEE347 pKa = 4.08PVPHH351 pKa = 6.61TEE353 pKa = 3.76LL354 pKa = 5.19
MM1 pKa = 7.52PPIRR5 pKa = 11.84KK6 pKa = 9.29LDD8 pKa = 3.66TRR10 pKa = 11.84AKK12 pKa = 9.72RR13 pKa = 11.84AAALRR18 pKa = 11.84RR19 pKa = 11.84GQIRR23 pKa = 11.84APRR26 pKa = 11.84RR27 pKa = 11.84RR28 pKa = 11.84GAPFRR33 pKa = 11.84YY34 pKa = 8.55RR35 pKa = 11.84TGTKK39 pKa = 9.86RR40 pKa = 11.84GRR42 pKa = 11.84TTAARR47 pKa = 11.84RR48 pKa = 11.84NGNTIALTTLSRR60 pKa = 11.84KK61 pKa = 8.65QEE63 pKa = 4.04NLSISYY69 pKa = 10.75RR70 pKa = 11.84EE71 pKa = 3.73MLEE74 pKa = 4.01FNDD77 pKa = 3.69MGGDD81 pKa = 3.61NGSTPCLIRR90 pKa = 11.84VNLNNPVIGGATPTGPDD107 pKa = 3.39TIVSVVGNLKK117 pKa = 9.46TGATDD122 pKa = 3.33PTFIPHH128 pKa = 6.36SYY130 pKa = 9.33EE131 pKa = 3.8NKK133 pKa = 9.95RR134 pKa = 11.84NLSDD138 pKa = 3.66RR139 pKa = 11.84LAEE142 pKa = 3.95YY143 pKa = 10.37FSIYY147 pKa = 8.66RR148 pKa = 11.84TAIVTSAEE156 pKa = 4.19VTVVCTPKK164 pKa = 10.62LNQLNGMTGNNRR176 pKa = 11.84SIVPFTQNRR185 pKa = 11.84ASEE188 pKa = 4.36DD189 pKa = 3.53PAVAGQATYY198 pKa = 10.5LYY200 pKa = 9.73QHH202 pKa = 6.89NANAMPQVEE211 pKa = 4.64VWSIRR216 pKa = 11.84QQNQGQLTSAAAGSPPLEE234 pKa = 4.22TLKK237 pKa = 10.84QGIPGMRR244 pKa = 11.84MTRR247 pKa = 11.84LNVVPNSTKK256 pKa = 10.13GVTYY260 pKa = 10.74KK261 pKa = 10.09MSYY264 pKa = 8.16TPKK267 pKa = 10.56SMYY270 pKa = 10.55AISDD274 pKa = 3.21WKK276 pKa = 11.0DD277 pKa = 3.04NKK279 pKa = 10.45KK280 pKa = 10.0VLKK283 pKa = 10.3IFNNAVKK290 pKa = 10.84NPDD293 pKa = 3.11QKK295 pKa = 10.48EE296 pKa = 3.66AYY298 pKa = 8.76MYY300 pKa = 10.82VGIAGRR306 pKa = 11.84FQGLDD311 pKa = 3.45PVASLANMGLPHH323 pKa = 6.72FNVEE327 pKa = 4.19VKK329 pKa = 10.54VKK331 pKa = 10.45YY332 pKa = 10.38NINFSEE338 pKa = 4.94RR339 pKa = 11.84YY340 pKa = 10.02NIDD343 pKa = 3.3GNNEE347 pKa = 4.08PVPHH351 pKa = 6.61TEE353 pKa = 3.76LL354 pKa = 5.19
Molecular weight: 39.21 kDa
Isoelectric point according different methods:
Peptides (in silico digests for buttom-up proteomics)
Below you can find in silico digests of the whole proteome with Trypsin, Chymotrypsin, Trypsin+LysC, LysN, ArgC proteases suitable for different mass spec machines.| Try ESI |
 |
|---|
| ChTry ESI |
 |
|---|
| ArgC ESI |
 |
|---|
| LysN ESI |
 |
|---|
| TryLysC ESI |
 |
|---|
| Try MALDI |
 |
|---|
| ChTry MALDI |
 |
|---|
| ArgC MALDI |
 |
|---|
| LysN MALDI |
 |
|---|
| TryLysC MALDI |
 |
|---|
| Try LTQ |
 |
|---|
| ChTry LTQ |
 |
|---|
| ArgC LTQ |
 |
|---|
| LysN LTQ |
 |
|---|
| TryLysC LTQ |
 |
|---|
| Try MSlow |
 |
|---|
| ChTry MSlow |
 |
|---|
| ArgC MSlow |
 |
|---|
| LysN MSlow |
 |
|---|
| TryLysC MSlow |
 |
|---|
| Try MShigh |
 |
|---|
| ChTry MShigh |
 |
|---|
| ArgC MShigh |
 |
|---|
| LysN MShigh |
 |
|---|
| TryLysC MShigh |
 |
|---|
General Statistics
Number of major isoforms |
Number of additional isoforms |
Number of all proteins |
Number of amino acids |
Min. Seq. Length |
Max. Seq. Length |
Avg. Seq. Length |
Avg. Mol. Weight |
|---|---|---|---|---|---|---|---|
0 |
619 |
265 |
354 |
309.5 |
35.19 |
Amino acid frequency
Ala |
Cys |
Asp |
Glu |
Phe |
Gly |
His |
Ile |
Lys |
Leu |
|---|---|---|---|---|---|---|---|---|---|
6.462 ± 1.511 | 1.454 ± 0.776 |
5.654 ± 1.978 | 4.362 ± 0.356 |
3.069 ± 0.46 | 6.785 ± 0.489 |
2.262 ± 0.989 | 6.947 ± 1.626 |
6.624 ± 1.097 | 5.977 ± 0.454 |
Met |
Asn |
Gln |
Pro |
Arg |
Ser |
Thr |
Val |
Trp |
Tyr |
|---|---|---|---|---|---|---|---|---|---|
3.069 ± 0.033 | 6.462 ± 2.251 |
5.008 ± 1.547 | 4.2 ± 0.214 |
7.593 ± 0.277 | 5.008 ± 0.56 |
8.078 ± 0.1 | 5.331 ± 1.265 |
1.131 ± 0.494 | 4.523 ± 0.497 |
Most of the basic statistics you can see at this page can be downloaded from this CSV file
Proteome-pI is available under Creative Commons Attribution-NoDerivs license, for more details see here
| Reference: Kozlowski LP. Proteome-pI 2.0: Proteome Isoelectric Point Database Update. Nucleic Acids Res. 2021, doi: 10.1093/nar/gkab944 | Contact: Lukasz P. Kozlowski |
